Ebachour Week 9
Contents
Electronic Journal
gene_name=[ENTER GENE NAME HERE] gene_id=$(curl -L "http://www.uniprot.org/uniprot/?query=559292+$gene_name&sort=score" | grep -o -P '(?<=tbody><tr id=").*(?=\" class=" entry s)') curl -L "http://uniprot.org/uniprot/$gene_id.xml"
Performing the tests were pretty straight forward. I simply followed the given instructions and documented the results. The hard part of this week's assignment was trying to figure out how we could use Uniprot to retrieve the ID and then use it to perform a curl command to get the XML for a gene from its gene name. We started by noticing that when you search Uniprot for a gene name, it outputs a list of results that match the search. We then discovered that if we put the taxon id that Dondi provided us with within the search, the first row of the table would be the yeast version that applies to that gene name. So the first part of the first curl command performs a search using any gene name and the Yeast taxon id and then gets the code of the page.
We then decided that with this curl command, we would just need to grep the first id in the row, and we did that by very specifically sifting through the code by retrieving everything between two very specific pieces of code. We then had the gene id in the gene_id variable which we then used to search Uniprot and get the web service response for that gene name. Let it be noted that this method only works if the requested gene exists for Yeast, and definitely doesn't work if you were to type in rubbish in the search.
GRNsight Test Results
We were tasked with testing the viewport size based on the test structure given in this PDF.
Test 1: PASS
Test 2: PASS
Test 3: PASS
Test 4: PASS
Test 5: PASS
Test 6: PASS
Test 7: PASS
Test 8: PASS
Test 9: PASS
Test 10: PASS
Test 11: PASS
Test 12: PASS
Test 13: FAIL
Test 14: FAIL
Test 15: FAIL
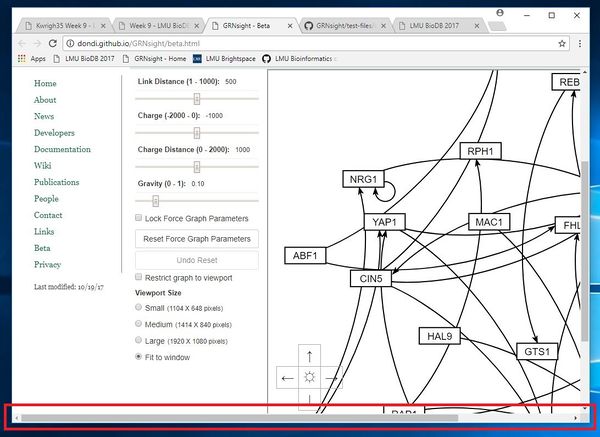
Test 13, 14, & 15 Fail Explanation: These three tests failed because the viewport doesn't truly "Fit to Window" when you get to smaller screen sizes. For example, when looking at the screenshot on the right, you would notice that there is a scrollbar on the bottom of the window. If you were to make the window just slightly larger, then that scrollbar would go away. This is because although the "Fit to Window" feature properly fits to window for most larger window sizes, but when the window gets below a specific width, it ceases to adhere to the full width of the window.
API
Upon determining how to go from a gene name/symbol to that gene’s individual data record (as shown in the Week 7 ajax-starter files), write up this process as a reproducible “recipe” in your electronic journal. In general, this recipe will consist of:
- The URLs to access in order to retrieve the desired data
- Any portions in these URLs that need to be substituted for specific queries, such as the gene name or ID within that web service
- Specific instructions on how to interpret the data returned by each URL so that you can extract exactly the information you need in order to proceed to the next step
UniProt
- Relevant documentation:
- Supplementary websites:
- Retrieve/ID mapping web page
- Technical information:
- You will encounter redirects in these web services; web browsers handle this automatically, but if using curl make sure to add the -L switch (i.e., curl -L …)
- Your URLs will include ampersands (&), which will need special handling with curl: in these cases, enclose the URL in apostrophes (e.g., curl -L 'http://www.uniport.org?query=this&type=that')
- UniProt primarily provides results in XML format; in one relevant step, the data can be provided in tab-delimited format, which might be easier to work with
- Miscellaneous information:
- You will encounter the need for a taxon ID, which identifies a specific organism; the taxon ID for our strain of S. cerevisiae is 559292
gene_name=[ENTER GENE NAME HERE] gene_id=$(curl -L "http://www.uniprot.org/uniprot/?query=559292+$gene_name&sort=score" | grep -o -P '(?<=tbody><tr id=").*(?=\" class=" entry s)') curl -L "http://uniprot.org/uniprot/$gene_id.xml"
Explanation above in the electronic journal
Acknowledgements
I worked with my homework partner Katie Wright in class. We also met face-to-face one time outside of class. We texted outside of class and met to work on the API part of teh assignment together.
While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Ebachour (talk) 21:24, 30 October 2017 (PDT)
References
- LMU BioDB 2017. (2017). Week 9. Retrieved October 27, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_9
Assignment Pages
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 14
- Week 15
Journal Entries
- Journal Week 2
- Journal Week 3
- Journal Week 4
- Journal Week 5
- Journal Week 6
- Journal Week 7
- Journal Week 8
- Journal Week 9
- Journal Week 10
- Journal Week 11
- Journal Week 12
- Journal Week 14
- Journal Week 15
Shared Journal Entries
- Shared Journal Week 1
- Shared Journal Week 2
- Shared Journal Week 3
- Shared Journal Week 4
- Shared Journal Week 5
- Shared Journal Week 6
- Shared Journal Week 7
- Shared Journal Week 8
- Shared Journal Week 9
- Shared Journal Week 10
- Shared Journal Week 11
- Shared Journal Week 12
- Shared Journal Week 14
- Shared Journal Week 15