Lenaolufson Week 9
Contents
- 1 10/27/15 Notes from class
- 2 10/29/15 Notes from class
- 2.1 TallyEngine
- 2.2 Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- 2.3 Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
- 2.4 OriginalRowCounts Comparison
- 2.5 Visual Inspection
- 2.6 .gdb Use in GenMAPP
- 2.7 Compare Gene Database to Outside Resource
10/27/15 Notes from class
- Go OBO-XMl and Uniport XML are linked by GOA and together their input is sent to the XMlpipedb software. GenMAPP builder converts the Prostgresac intermediate database to a form where it can be used to create GenMAPP Gene Database .gdb, which is then used to analyze microarray data, that came from downloading and analyzing microarray data, with the GenMAPP program.
- 13-Oct-2015 07:31 time the database put in the updated file: 46.V_cholerae_ATCC_39315.goa. I accessed and downloaded the file on 10/27/15
- Vcholerae_2015_10_27_gmb3build5
- For exporting: -owner:Lena Olufson -species: Vibrio cholerae
- Used this page for instructions on how to perform an export of the Vibrio cholerae GenMAPP Gene Database: https://xmlpipedb.cs.lmu.edu/biodb/fall2015/index.php/Running_GenMAPP_Builder
Export Information
Version of GenMAPP Builder: gmb3build5
Computer on which export was run: HP LV2311-
Postgres Database name: Vcholerae_2015_10_27_gmb3build5
UniProt XML filename (give filename and upload and link to compressed file): Media:Uniprot-organism-243277.xml.gz
- UniProt XML version (The version information can be found at the UniProt News Page): 2015_10
- UniProt XML download link: http://www.uniprot.org/uniprot/?query=organism:243277
- Time taken to import: 2.92 minutes
- Note:
GO OBO-XML filename (give filename and upload and link to compressed file): Media:Go_daily-termdb.obo-xml.gz
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped):
- GO OBO-XML download link: http://geneontology.org/page/download-ontology#Legacy_Downloads
- Time taken to import: 7.23 minutes
- Time taken to process: 4.32 minutes
- Note:
GOA filename (give filename and upload and link to compressed file): Media:46.V_cholerae_ATCC_39315.goa
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site):
- GOA download link: http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa
- Time taken to import: 0.07 minutes
- Note:
Name of .gdb file (give filename and upload and link to compressed file): Media:Vc-Std_20151027_LO.gdb
- Time taken to export: 1 hour and 19 minutes
- Start time: 3:53:53 PM PDT
- End time: 5:12:03 PM PDT
Note: This lasted longer than the class period so I left the computer to continue the export while I was out of the classroom, I left a note on the computer to please not close any windows that are open and running on this computer.
10/29/15 Notes from class
- Uniprot is an xml file, Go is an xml file, COA is a tab delimited file and these three files get imported into GenMAPP builder which exports files as a gene database. GenMAPP builder also outputs to Postgre SQL.
- There are 7664 entries in the OrderedLocusNames table and the OriginalRowCounts, some have VCA some have VC some have underscores and some do not. The ID's in both the xml and PostgresDB tallied to 3831, which is roughly half the amount of IDs than in the MS Access tally.
TallyEngine
- Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases.
- Choose the menu item Tallies > Run XML and Database Tallies for UniProt and GO...
- Take a screenshot of the results. Upload the image to the wiki and display it on this page.
- For more information, see this page.
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Follow the instructions found on this page to run XMLPipeDB match.
Are your results the same as you got for the TallyEngine? Why or why not?
- I used the command:
java - jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xmlto generate back the total unique matches are 3831, which is the same number of gene IDs as the TallyEngine outputted.
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
For more information, see this page.
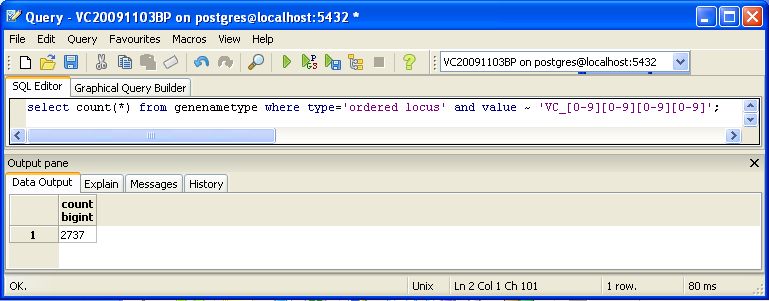
You can also look for counts at the SQL level, using some variation of a select count(*) query. This requires some knowledge of which table received what data. Here’s an initial tip: the gene/name tags in the XML file land in the genenametype table. A query on this table counting values from this table that were marked as ordered locus in the XML file matching the pattern VC_[0-9][0-9][0-9][0-9] would look like this:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';
In pgAdmin III, you can issue these queries by clicking on the pencil/SQL icon in the toolbar, typing the query into the SQL Editor tab, then clicking on the green triangular Play button to run.
Are your results the same as reported by the TallyEngine? Why or why not?
- When using the command provided above, the result is 2737 gene ID's. This decrease in number is due to the lack of accounting for the ID's with VC_A....
- By using the command:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9]'the result is 3831 gene ID's and therefore is the same as the previous XMLPipeDB Match and TallyEngine results found.- This command worked because it A> in so that pgAdmin III could include gene ID's that either do or do not have an A proceeding the underscore.
OriginalRowCounts Comparison
Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
Benchmark .gdb file: http://sourceforge.net/projects/xmlpipedb/files/V.%20cholerae%20Gene%20Database/V.%20cholerae%2020101022/Vc-Std_External_20101022.zip/download
Copy the OriginalRowCounts table from the benchmark and new gdb and paste them here: https://xmlpipedb.cs.lmu.edu/biodb/fall2015/images/c/ca/OriginalRowCounts.pdf (found by classmate)
Note: I was having trouble finding the original row counts and table on my own so I looked at one of my classmate's assignment page and used his link.
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
- No, there are a large number of dates missing from the system, around 20.
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
- It depends on which tables; UniProt all appear to be in the same format, RefSeq the IDs are not all the same because some have underscores and some do not, and OrderedLocusNames yes all the IDs appear to be in the same format.
Note:
.gdb Use in GenMAPP
Note: I was able to download my .gdb file from this wiki page and select it as the gene database for the GenMAPP program without difficulty and it took hardly any time at all.
Putting a gene on the MAPP using the GeneFinder window
- Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
Note:This step was unable to be performed due to class error.
Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
Note: I was able to use the previous Merrell et. al spreadsheet to test the new gdb file I had used as the database. However, I did not run the ID check and do not know the exact numbers.
Coloring a MAPP with expression data
Note: I was able to color a map as shown in class for the previous assignment just fine and it all seemed to work well and smoothly. It did not take very long at all.
Running MAPPFinder
Note: I was able to run MAPPFinder with my imported database and the hierarchy of GO was visible on my computer screen. The different colors were all present and displayed and the same sorting and filters worked as before when navigating through the list.
Compare Gene Database to Outside Resource
The OrderedLocusNames IDs in the exported Gene Database are derived from the UniProt XML. It is a good idea to check your list of OrderedLocusNames IDs to see how complete it is using the original source of the data (the sequencing organization, the MOD, etc.) Because UniProt is a protein database, it does not reference any non-protein genome features such as genes that code for functional RNAs, centromeres, telomeres, etc.
Note:
Loyola Marymount University: website
| Weekly Assignments | Individual Journal Pages | Shared Journal Pages |
|---|---|---|
|
|
|
|