Introduction to the Command Line
- For much of the computer work in this course, we'll be using a computer interaction style that's very different from what's familiar to most of us: the command line.
- The reason for this is that the command line actually offers a surprising wealth of text processing tools, some of which are more powerful than the applications that we're accustomed to using.
- It is clear, however, that one needs a lot more "up-front" training with the command line, and that's what this page hopes to provide.
- To put things in context—as can be seen from the videos below, everything "old" will be new again: using the command line is ultimately a lot like "talking" to science fiction computers, but with typing and reading instead of talking and listening:
Contents
Command Line Basics
Working with a command line is a cycle (i.e., a loop) of:
- The computer indicating that it is ready for the next command (via a prompt)
- The user (you) typing in a command
- While typing a command, you may use the arrow, backspace, and other keys to edit what you've typed so far
- To completely start over, hold down the control key then hit c (i.e., "control-c"); you'll go back to a fresh prompt
- When you're ready with your command, hit the Enter or Return key
- Many users experience some fear with typing a command—this is understandable, but rest assured:
- Typically, the worst thing that happens is that the computer did not "understand" the command, thus doing nothing
- While there are harmful commands, (a) the likelihood of your typing one at random is very low, and (b) most modern operating systems (including the one on the Keck Lab workstations) will prevent you from doing the really dangerous stuff anyway
- The computer performing the command, then showing you the result of that command
- "Rinse and repeat"
If all goes well, then with each command you type, you get closer to accomplishing your goal.
The Command History
To assist with this command entry cycle, modern command lines keeps track of the commands you type—essentially, a command history. This history shows up in a number of ways:
- If you press the up or down arrow keys at the command prompt, you will move back and forth through your history. If you see the command you'd like to perform, press Enter or Return, and the computer will try to perform that command
- You can do "variations" on past commands by immediately editing what shows up; that is, you can press the up or down arrow keys until you see a command that's similar to the one you want to do, then use the left/right arrow keys, backspace, and other keys to edit it
- The control-c shortcut is always there if you want to start over
“Autocomplete” with the Tab Key
While working on the command line, the Tab key (the one to the left of the Q key) provides a convenient “autocomplete” function. No matter where you are in your command, it can be a good idea to tap the Tab key if you're in the middle of typing something out.
- If the computer concludes that what you've typed in so far can mean only one thing, then it will “autocomplete” what you have by spelling everything out.
- If the computer concludes that what you've typed in so far can still mean multiple choices, the first Tab will do nothing (though on some systems you may hear a beep); hitting Tab a second time will show you the computer’s “guesses” as to what you might be typing.
- You can keep typing a few letters, then hitting Tab, until there is only one choice and the computer spells everything out for you. We’ll see instances of this later, in terms of the other commands.
In general, it's a good idea to periodically type the Tab key, either when you can only remember the first few letters of a command or file name, or if you want to save some typing and know that you've typed in enough letters so that only one choice is available.
Your First Command: exit
The first command to try is:
exit
Typing this command ends your "command session" with the computer. Most of the time, this closes the window into which you've been typing your commands (typically called a "terminal"). Sometimes, you get a message that your session is finished, but you still need to close the window manually. In any case, exit means you're done, and you can quit whatever program you were using to get to the command line (e.g., Terminal, PuTTY, etc.).
- Since we just mentioned the Tab key, try this—type only the first two letters of exit:
ex
- Now hit the Tab key twice. You should see a list of command that start with ex, similar to this:
ex exo-csource export extractattr exchangewizard exo-desktop-item-edit expr extractkmdr exec exo-open extcheck extractrc exif.py exo-preferred-applications extend_dmalloc extractres exifautotran expand extensionproxy exit expiry extract_a52
- Now type the letter i:
exi
- If you hit the Tab key twice again, you’ll see that the list has narrowed down to this:
exif.py exifautotran exit
Of course, this might not be useful for a command as short as exit, but in any case this helps demonstrate the Tab key’s functionality.
Files and Folders on the Command Line
Before we move on into actual text processing commands, let's look at some key concepts and commands for just "getting around" the files on a computer using the command line.
How Files and Folders Look on a Command Line
Just like with the computers we use everyday, we can access our files and folders on the command line. Unlike the computers that we use everyday, we don't see any icons, folders, or pictures; instead, command lines represent files and folders as text expressions called paths.
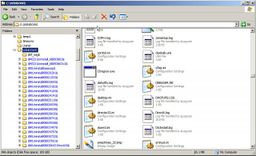
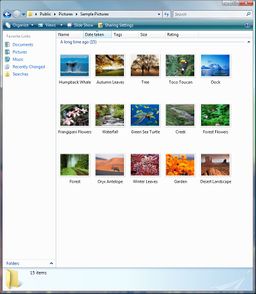
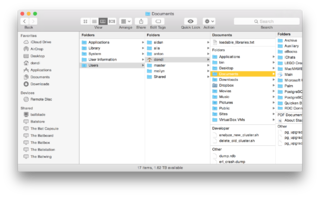
You've probably seen displays like the ones below on your computer. Click on them to see them full-sized:
On a command line, the folders displayed by the windows above are expressed, respectively, as:
/WINDOWS /Users/Public/Pictures/Sample Pictures /Users/dondi/Documents
(Windows Vista sometimes hides the Users folder from the window, and that's why you don't see Users in the screenshot)
The way to read a path is to separate out the slash (/) characters; starting from the left, each slash marks off a folder that is inside another one. The very last item after the last slash represents the actual file or folder that is indicated by the path. Thus, you would interpret a path like:
/home/xmlpipedb/public_html
To actually mean the public_html file or folder inside the xmlpipedb folder inside the home folder, which in turn sits at the "top" or "root" of the file system—that is, it's a folder that isn't inside any other folder. As a tree, the path above looks like this:
/ (root)
|
+-home
|
+-xmlpipedb
|
+-public_html
- If there is a folder called data within the folder above, its path would be:
/home/xmlpipedb/public_html/data
- If there is a file called prokaryote.txt within the above data folder, then its path would be:
/home/xmlpipedb/public_html/data/prokaryote.txt
Your “Working” Directory: pwd
On everyday computers, we frequently see a window—typically when we're trying to save our work or browse our desktop—that gives us a concept of "where" we are among all of our files. While we don't have such a window on the command line, the computer does keep track of what folder we're in. This folder is called the working directory.
- Terminology alert: The word directory is a synonym for what everyday computers call a folder. They mean the same thing—a folder or directory is an entity on the computer that keeps files, some of which may themselves be more folders or directories. For this write-up, folder and directory will be used interchangeably, to get you accustomed to seeing both words.
If, at any time, you forget "where" you are in your files, this command will display your working directory:
pwd
You'll notice that, because commands involve a lot of typing, they tend to be brief or even abbreviated. While this makes them harder to remember, they still resemble what they mean to do; for example, pwd stands for print working directory—which is exactly what that command does.
Your “Home” Directory
When you connect to a command line for the first time, the working directory typically starts at your user account's home directory. This is the folder in which your user account is allowed to create files and otherwise do all kinds of other work. Most computers today have this concept; on Windows computers, your home directory is typically displayed as a folder icon with a name like Joe's Documents. On Mac OS X computers, the home directory is typically displayed as a house icon with the same name as your login.
If you invoke pwd immediately upon logging into a Keck lab computer, you will be in your home directory within that system. Dr. Dionisio's home directory on my.cs.lmu.edu, for example, is:
/nfs/home/dondi
Fortunately, there is a shorthand for any user's home directory in the Keck lab, which we will see and use in the next section.
- Terminology alert: The word invoke is commonly used to mean “make the computer perform a command,” in this case by typing the command and hitting the enter or return key.
Getting Around
On everyday computers, you move from folder to folder by clicking or double-clicking the folder icon that you like. Some systems have a "folder up" button that you can use to move to the folder that contains your current folder.
The command line has similar commands, with the difference being that they are typed instead of invoked with a mouse.
List Files: ls -F
To see the files inside your current folder (i.e., your working directory), type this:
ls -F
Here, think of ls -F as “list files.” While the exact contents of your home directory may vary, ls -F typically produces something like this:
Desktop/ Downloads/ Pictures/ public_html/ Templates/ Documents/ Music/ Public/ sandbox/ Videos/
Each name is a file inside your folder—so it's really a lot like the screenshots shown before, but without icons. Since we don't have icons, how can we tell if a name represents another folder? That is indicated by the slash (/) at the end of the name. In the listing above, public_dav and public_html are folders.
Change Directory: cd
Let's say that you want to change your working directory to something else. The command for this is:
cd <new directory>
To remember this, cd stands for change directory. The <new directory> part in the command above is the name of the directory that you want to change to. Thus, if you want to “go” to the public_html directory/folder shown previously, you would type:
cd public_html
After doing this, type:
pwd
...to convince yourself that you did successfully go to the new directory; type:
ls -F
...to see what's in there (probably nothing, if you have a relatively new Keck lab account).
What about that "folder up" button? That is indicated by the special name .. (two periods one after the other). To “go up” a directory, type:
cd ..
If you then type:
pwd
You should be back in the folder that contained public_html.
A few shortcuts are available when the directory you want is someone (or your own) home directory.
- If you type cd by itself:
cd
...you will “teleport” back to your home directory, no matter where you are. - If you type cd followed by ~<username>, where you substitute <username> for another account in the system:
cd ~dondi
...you will “teleport” to that user’s home directory, no matter where you are. ~<username> (that first “squiggly” symbol is called a tilde) is the aforementioned shorthand for expressing a user’s home directory.
The Tab Key and cd
Recall that the Tab key offers an “autocomplete”-like feature on the command line. This totally works with the cd command. For example, if your home directory has folders named Documents and Downloads (and those are the only folders that begin with a capital “D”), typing this while on your home directory:
cd D
...then hitting Tab, will make the system automatically extend what you typed to:
cd Do
That’s because the only two choices that begin with D actually also begin with Do, so the system is “sure” that what you want at least starts with that. As before, if you hit Tab one more time, you’ll get:
Documents/ Downloads/
...since those are the actual possible choices for cd.
So remember, hit Tab early and often! :)
- Note: The Tab key also works with ls -F, but we’ll skip over that for now.
Basic File Commands
There are lots of file commands, but for introductory purposes we’ll present the two basic ones which you have probably used in a non-command way already.
Copy File: cp
To copy a file from one place to another, use this command:
cp <file to copy> <destination of copy>
You have probably copied files before, such as from a laptop to a flash drive. In mouse/touch environments, this operation is typically a drag-and-drop—you hold down a mouse or trackpad on the icon of the file to copy, then drag it to the icon of the flash drive destination. Usually, the mouse cursor changes to show that you are about to perform a file copy (a common sign is the appearance of a “+”). In addition, there is a safety net that warns you if you are about to copy a file into a destination where a file of the same name already exists.
In Windows, you can also click on an icon to select the file, choose Copy from a menu, then navigate to the destination and finally click Paste. Note the similarities in the pattern, regardless of the specific mechanism: indicate the file to copy, then indicate where the copy should go. If you think of the cp command in this way, that may make the learning go more smoothly.
In the end, though, there are some differences to note:
- Already mentioned is the way we are used to receiving warnings if we are about to copy over a file that already exists. The command line does not do this by default. If you would like to play it safer and be given that warning, ad a -i to the command, separated by spaces:
cp -i <file to copy> <destination of copy>
- The “i” stands for “interactive,” which may make it easier to remember.
- Don’t forget, you can use various commands in any order. You can use ls or cd at any time to get a handle on what files are around and where. After performing a copy, you can use those commands again to make sure that the file really did get copied. Part of getting used to the command line is the ability to string individual commands together in a meaningful way.
- We are accustomed to copying files to a folder or directory, and indeed cp can work that way. However, it does have one other option which might not immediately be obvious in other user interface styles: you can copy a file and give the copy a new name in a single command. For example, if you want to copy a file called genetic-code.sed from the ~dondi/xmlpipedb/data folder into your home directory but want to rename the copy as gc.sed, you can do this in one line:
cp ~dondi/xmlpipedb/data/genetic-code.sed ~/gc.sed
- (remember the ~ shortcut for home directories above)
- Finally, the .. shorthand for the folder “above” the current one still holds here. That is another pattern to realize about the command line: the “vocabulary” of shorthand and symbols typically apply across whole families of commands. Thus, once you learn how to use one command well, chances are that you are a leg up in learning other related ones.
Move File: mv
Sometimes you don’t want to copy a file, but just move an existing one from one folder to another. The command for this is mv (“move”), and its structure is very much like that of cp:
mv <file to move> <new location of the file>
Note again the conceptual similarity between this command and what you may be used to (i.e., drag-and-drop of a file icon from one place to another; selecting a file, choosing Cut from a menu, then choosing Paste at the file’s new location): all commands indicate the file to move, then the destination of the file. It’s just how you express this that differs for each mechanism.
Most of the bullet points for cp also apply to mv, including the -i safety net option (remember what we said about having that shared “vocabulary?”). Plus, mv has one last twist of its own...
Rename File: mv (!)
Yes, the command to rename a file is also mv. That’s because the command line does not distinguish between a move and a rename—renaming a file is simply “moving” it from a file of one name to a file of another. Thus, renaming a file is:
mv <old name of file> <new name of file>
Note how this reflects a certain minimalism or non-redundancy in how commands are defined—instead of creating a whole new command for some operation, if another command effectively does the same thing, then the choice is to use that instead of defining another one.
Processing Text
Most of what you’ll need to do on the command line initially is to process text. That is, you’ll be manipulating text data, such as:
gaatccattcagc
Note that this nucleotide sequence, when represented digitially, is simply a sequence consisting of the letters a, c, g, and t.
The computer distinguishes between lowercase and uppercase letters, so the text above is not the same as:
GAATCCATTCAGC
When the sequence of letters is stored on a computer’s disk (and not just typed out), then it is given a name and becomes a text file. Text files can also have multiple lines, such as:
GGCCCTCAGGCAAGGGCTCTGAAGTCAGGGTCACCTACTTGCCAGGGCCGATCTTGGTGCCATCCAGGGG GCCTCTACAAGGATAATCTGACCTGCAGGGTCGAGGAGTTGACGGTGCTGAGTTCCCTGCACTCTCAGTA GGGACAGGCCCTATGCTGCCACCTGTACATGCTATCTGAAGGACAGCCTCCAGGGCACACAGAGGATGGT ATTTACACATGCACACATGGCTACTGATGGGGCAAGCACTTCACAACCCCTCATGATCACGTGCAGCAGA CAATGTGGCCTCTGCAGAGGGGGAACGGAGACCGGAGGCTGAGACTGGCAAGGCTGGACCTGAGTGTCGT
Sometimes, text is readable in a certain way. For example, many gene sequences are stored with a preliminary line, describing the sequence:
>ref|NT_011255.14|Hs19_11412:1-7286004 Homo sapiens chromosome 19 genomic contig, reference assembly
Such information is somewhat readable by us, but does take a little additional knowledge to understand it fully.
Basic Text Commands
On everyday computers, we typically work with files using a mouse; we start by finding its icon, then we double-click on it. When we double-click on the file’s icon, a window usually appears bearing the content of that file. Within that window, you can invoke menu items for finding or replacing text. If we want to change the text in our own way, we start typing into the window. Finally, if we want to make our changes permanent, we save the file.
In this section, we will look at command line equivalents for all of those operations, except for saving. You won’t need it for now, and there are so many variations to saving your modifications that it’s better to delay that for another time.
Location of Sample Files
Most of your initial work involves files that are currently stored in the data folder within the xmlpipedb directory of the dondi user account. So let’s go there using commands that you’ve already learned. You start working inside the data folder by invoking cd:
cd ~dondi/xmlpipedb/data
(yes, the Tab key works here—feel free to experiment a bit and see how much typing you can save)
You can verify that you are indeed in this folder by typing:
pwd
...which should display:
/nfs/home/dondi/xmlpipedb/data
To look at the files available here, type:
ls -F
...and you should see something very close to:
18.E_coli_MG1655.goa genetic-code.sed infA-E.coli-K12.txt prokaryote.txt 493.P_falciparum.xml hs_ref_GRCh37_chr19.fa movie_titles.txt xmlpipedb-match-1.1.1.jar
There are eight (8) files in this folder, each with different types of content. The remainder of this section assumes that your working directory is /nfs/home/dondi/xmlpipedb/data.
Viewing: cat
Since command line interaction does not have floating, separate windows, viewing a file has to happen within the command line window. There are two command to do this; the first one, cat, is the simplest way to display a file:
cat <filename>
You replace <filename> with the name of the file that you’d like to display. For example, try this:
cat prokaryote.txt
Invoking this command should give you this:
tctactatatttcaataggtacgatggccaaagaagacaatattgaacttgaaacgttgcctaataccatgttccgcgtataacccagccgccagttccgctggcggcattttaac
Now try this:
cat genetic-code.sed
What happened? You do get shown the contents of the genetic-code.sed file. But, unless your command line window was huge, you may have noticed that the content of this file ran off the screen, forcing you to scroll up and down to see the whole thing.
Merely enlarging the command line window won’t necessarily help: one can always come up with a text file that is bigger than the command line window. For example, try this:
cat movie_titles.txt
Huge, isn’t it? Give it a few seconds to get displayed in its entirety.
- As you might have guessed, Tab autocompletion works with cat. You can type:
cat gen
...and the system autocompletes genetic-code.sed, since that is the only file in the working directory that begins with gen. - In case you were wondering, cat is short for concatenate. The reason for this name actually has to do with other things that you can do with cat, which we will skip for now.
“Paging:” more
Fortunately, there is a command that takes the size of the command line window into account, and will allow you to look at a text file a screenful at a time. This command is more (as in, it waits for you before it displays “more” information). It takes the same information as cat; namely, a filename:
more <filename>
Try this:
more movie_titles.txt
What happens now? You see a command line window’s worth of the file, and that’s all. Note how the bottom of the command line window says:
--More (0%)--
This indicates that you haven’t seen the entire file yet, and that you’re around 0% of the way down.
From here, pressing some keys allows you to move around the file:
space bar Moves forward a command line window’s worth at a time enter or return Moves forward a line at a time b When viewing files, moves backward a command line window’s worth at a time q Quits from more
Practice moving back and forth through the movie_titles.txt file to get the hang of it. When you’re done, type q to get out of more.
- After you quit from more, don’t forget that the up and down arrow keys allow you to look at previously-typed commands. Thus, if you want to look at the file again, don’t bother typing the whole command over—just hit the up arrow key, then press enter or return, and you’re back in more, paging through that file.
- more actually has quite a few, well, more features while paging through a file. These will have to wait for another time.
- more is what is called a paging program, since it allows you to “page” through large files. There is actually an alternative pager, called less, so named as a play on the saying “Less is more.”
Counting: wc
Everyday applications like Microsoft Word can count the number of words, characters, and other units within the documents that they edit. The command line has an equivalent for this too: wc, short for word count:
wc <filename>
Try this:
wc movie_titles.txt
You should see this:
17770 65246 577547 movie_titles.txt
Not as pretty as Microsoft Word, but just as useful: the first number is number of lines, the second number is the number of words, and the last number is the number of characters (essentially the file size) in movie_titles.txt. Lines, words, characters.
Counting Multiple Files
Here’s something that you can do with wc that’s a little harder with Microsoft Word’s counting feature. Invoke this command:
wc *
What shows up? It should look a lot like this:
43640 716372 7886582 18.E_coli_MG1655.goa
1118618 3025873 38244964 493.P_falciparum.xml
64 64 640 genetic-code.sed
801592 801654 56913098 hs_ref_GRCh37_chr19.fa
1 1 593 infA-E.coli-K12.txt
17770 65246 577547 movie_titles.txt
1 1 117 prokaryote.txt
35 89 4147 xmlpipedb-match-1.1.1.jar
1981721 4609300 103627688 total
Yes, wc can handle multiple files, and when it can tell that you are trying to count multiple files, it even totals everything up for you. The asterisk (*) after wc is called a wildcard, meaning that it can match any filename in the working directory.
This is a first hint at the additional power that can be at your fingertips once you get used to working on the command line.
Lines Add a Character
Let’s break down the wc output for the file genetic-code.sed:
wc genetic-code.sed
This displays:
64 64 640 genetic-code.sed
So, 64 lines, 64 “words,” and 640 characters, right? But if you cat or more genetic-code.sed, you’ll see lines like these:
s/gua/V/g s/gug/V/g s/ucu/S/g s/ucc/S/g
If you count the characters, that’s just nine (9) per line! But if there are 64 lines in this file, and 640 characters overall, then shouldn’t there be ten (10) characters per line?
The answer is yes, and the reason that you only see 9 characters per line is because the 10th character is invisible—it is the end of the line itself. Technically called a “newline,” it is an invisible symbol that tells the computer to go to the next line of text. Newlines are written with a backslash followed by a lowercase n:
\n
Thus, each line of the genetic-code.sed file actually looks, to the computer, like:
s/gua/V/g\n s/gug/V/g\n s/ucu/S/g\n s/ucc/S/g\n
If you think of \n as a single character, then that does result in 10 characters per line, and 64 lines of that adds up to 640 characters total.
Remembering to count the newline is crucial when working with data such as gene sequences. For example, if you do this:
wc prokaryote.txt
...you’ll see this:
1 1 117 prokaryote.txt
That’s one line, one “word” (i.e., the entire sequence), and 117 characters. But since one of those characters is the newline, then this file really only has 116 nucleotides.
You’ll need to take this into consideration with some of the journal questions.
Finding: grep
Now we’ll start looking at some seriously powerful commands. The first one has to do with finding specific text within a file. The command that does this is called grep. Unfortunately, explaining the name is too complicated for now, so let’s just leave it at grep:
grep "<pattern>" <filename>
Note how, this time, grep needs two pieces of information: a “pattern” to find, and the file(s) within which to find that pattern. Like wc, you can replace <filename> with a wildcard:
grep <pattern> *
This will try to find the desired pattern in every file in the working directory.
Try this:
grep "Romance" *
(remember that we’re assuming that the working directory is ~dondi/xmlpipedb/data on a Keck lab computer) You should see something like this:
movie_titles.txt:979,1990,A Moment of Romance movie_titles.txt:3082,1985,Murphy's Romance movie_titles.txt:4038,1982,A Fine Romance: Set 2 movie_titles.txt:4805,1993,True Romance movie_titles.txt:6175,1981,A Fine Romance: Set 1 movie_titles.txt:9103,1914,Tillie's Punctured Romance movie_titles.txt:9113,1979,A Little Romance movie_titles.txt:9331,1999,Romance movie_titles.txt:9535,1941,Irene Dunne Romance Classics: Love Affair/ Penny Serenade movie_titles.txt:13760,1983,A Fine Romance: Set 3
grep’s output states that it found “Romance” multiple times in the file movie_titles.txt, and nothing else. After the filename, it displays a colon (:) then prints the actual line that had the pattern.
Because movie_titles.txt is a file containing thousands of movies, grep "Romance" * effectively gives you a list of all movies with “Romance” in their titles. If you choose your pattern well, you can find all kinds of other information. Try this:
grep "1914" movie_titles.txt
Did you think that this will give you a list of movies released in 1914? Not exactly:
1914,2002,Damaged Care 9103,1914,Tillie's Punctured Romance 10898,1914,Cabiria 11914,2001,The Blue Planet: Seas of Life: Tidal Seas - Coasts
Remember that grep is a pure pattern matcher: it looks only for text matches, and is not aware of what that text means. Thus, it found “1914” not only in the release year, but also in the numerical ID at the beginning of the line.
We can observe that, in this file, the release year is preceded by commas (,). Thus, we can do this:
grep ",1914," movie_titles.txt
...and now we get the desired result:
9103,1914,Tillie's Punctured Romance 10898,1914,Cabiria
Non-Exact Matches
Exact matches are interesting, but most other everyday applications can do this without a problem. Note how we said that grep can match a pattern and not just search text. It turns out that grep can “understand” a wide variety of symbols that represent different patterns of text.
A period (.) represents any single character. Thus, this pattern:
grep "St..r" movie_titles.txt
...produces all lines that have “St” and “r” with any two symbols in between:
252,2002,Stuart Little 2 462,2005,Classic Cartoon Favorites: Starring Donald 2017,1991,The People Under the Stairs 2018,2005,Classic Cartoon Favorites: Starring Goofy 4906,1999,Stuart Little 7021,1995,Stuart Saves His Family 7699,2004,Bratz: The Video: Starrin' & Stylin' 8436,2005,Classic Cartoon Favorites: Starring Mickey 12016,2005,Classic Cartoon Favorites: Starring Chip 'n' Dale 13099,1946,The Spiral Staircase 13203,2003,And Starring Pancho Villa as Himself 14448,1998,The Staircase 16076,2003,Wishing Stairs
Let’s change the text file for now. Since grep only cares about text, and not what that text means, you can use patterns that represent information other than movie titles...such as nucleotide sequences. Try this:
grep "TATA...ATG...ATT...TGA" hs_ref_GRCh37_chr19.fa
This displays any lines with the sequences TATA, ATG, ATT, and TGA, with exactly three nucleotides in between. For what it’s worth, this is what you get on the sample file:
AGAGCAGTGGTATGCACTGCTCTATTGAGTATACTTATGGTTATTTATTGATTATATGCTAAATAAGGGG CTAACTATACAAATGGAAATTTTATGATGATAAGATCAGTAGTTACTGAGATATTTATGTGACAAGCTGA TGTCTCCCTTTCAGTGTGGGGGAGCTCACTATAGCCATGGAGATTCCATGAAACATTTTAGCACCAAACA
That’s three lines out of how many total? Hope that gives you an idea of how much time grep can save you, once you learn how to use it.
Here are some other patterns that you’ll find useful. Needless to say, this is just the tip of the iceberg; as you get more comfortable with grep, you can learn more and more variations for text patterns.
[<characters>] Matches lines that have any of the characters listed in <characters> ^pattern Matches lines that start with the given pattern pattern$ Matches lines that end with the given pattern pattern* Matches zero or more repetitions of the given pattern
As mentioned, there are many more, but this is a start.
A final useful feature is negation. When you ask grep to negate a pattern, it will display lines that don’t match the pattern. This is done by inserting a -v in the command:
grep -v [AEIOUaeiou] movie_titles.txt
As stated above, the brackets [ ] match any of the characters in between. Having -v makes grep find the lines that do not have A, E, I, O, U, a, e, i, o, or u at all. The line above effectively gives you movie titles that don’t have any vowels.
- Recall that computers distinguish uppercase and lowercase letters, so it’s necessary to include both if you want to wipe out all vowels. What do you get if you remove the uppercase versions? The lowercase versions?
Putting Commands Together
There are a few more commands left to cover, but for now we have enough to look at perhaps one of the most powerful aspects of a command line interface: putting multiple commands together. This ability lets you let one command work off the results of another command, producing a classic example of “the whole is greater than the sum of its parts.”
The “Anatomy” of a Command
Think of a command as a box, with text coming in one side, followed by text coming out another:

|
Well, text is text, right? What’s stopping us from using the output text of one command and making it the input text of another one? The answer is, nothing. This can be done:

|
If you think about the text data “flowing” from one command to another, with each command performing some processing on each successive stream of text, you’ll see the origin of the word for this activity: piping. We are essentially forming a pipe through which text “flows,” and at each stop, the text changes a little bit, depending on what the specific command does.
Creating a Pipe on the Command Line
A single character is responsible for creating a pipe: |. We typically call this the “vertical bar,” but in this context, it is called the “pipe character.” Let’s start with an example.
At this point, you’ll probably recognize this command as finding all lines with the word “Leonardo” in it:
grep "Leonardo" movie_titles.txt
This will yield:
7410,1997,Leonardo da Vinci: Renaissance Master 7564,1972,The Life of Leonardo Da Vinci 14084,2000,Leonardo DiCaprio: Double Feature
Now, see what happens when you pipe this text to wc:
grep "Leonardo" movie_titles.txt | wc
Notice how, instead of giving wc a filename, the | character denotes that it should use whatever grep "Leonardo" movie_titles.txt produces. Here’s the result:
3 15 133
...3 lines, 15 words, 133 characters. Since movie_titles.txt lists one movie per line, we have effectively written a command sequence that tells us how many movies in that file have “Leonardo” in its title.
That’s pretty much all there is to it—if you think of the output of one command as being a possible input for another, just stick a | in between the two commands, without giving a filename to the latter commands in the pipe.
Of course, you don’t have to stop at two. If you want to count how many movies were released in 2000 that don’t start with “The”, then you can do this:
grep ",2000," movie_titles.txt | grep -v ",The " | wc
...which yields:
1056 3703 33889
Can you somehow check your work? With a few other commands, you can. First you can count the movies released in 2000 that do start with “The”:
grep ",2000," movie_titles.txt | grep ",The " | wc
(no -v eliminates the negation of the pattern) This displays:
178 708 5798
Finally, let’s just count the movies that were released in 2000:
grep ",2000," movie_titles.txt | wc
This produces:
1234 4411 39687
Adding up the lines, we find that 1056 + 178 = 1234. So the numbers are self-consistent; these pipes work.
Pipes Without Files: Just Type Directly
You might have noticed that, when we want the input text to come from another command instead of from a file, you simply drop the filename from the command that’s in the pipe. In the previous examples, though, the very first command does have a filename—after all, you have to get your initial text from somewhere, right?
Interestingly, you don’t need a filename, even for the very first command. It turns out that, for these text processing commands, if you don’t give them a filename, then they just use whatever you type in after invoking the command. Try this; first, type the command; you’ll notice that just doing this does not get you back to a new command prompt. Instead, the computer seems to just sit there. In reality, it’s waiting for data, from you:
wc
Into this waiting void, type the following, hitting enter or return to go to the next line:
The quick brown fox jumps over the lazy dog. That's all, folks!
After the second line, hold down the control key, then tap the d key (i.e., “control-d”). This tells the command that you’re at the end of your input. This is what shows up, followed by the command line prompt:
2 12 64
Two lines, 12 words, 64 characters—exactly what you typed in. Thus, even files themselves can be replaced with your own live input. This feature is great for experimentation and trying things out. For example, if you aren’t sure what the "[qz]" will match with grep, you can just type in that command without a filename:
grep "[qz]"
Then, you can type a variety of lines, to see whether or not they match. If a line matches the pattern, grep will repeat it; if not, then it won’t. Type control-d on a blank line to tell grep that there’s no more text to process, and you’ll be back on the command line.
For example, here’s what appears on the screen if the user types “hello world,” “quit bugging me,” “Quit bugging me,” “what's up,” “Zounds!,” “zoundz!,” then control-d:
hello world quit bugging me quit bugging me Quit bugging me what's up Zounds! zoundz! zoundz!
Since only “quit bugging me” and “zoundz!” match the [qz] pattern, then only those lines are repeated by grep.
A Few More Commands
Now that you know about pipes and direct input, we can look at a few more commands that benefit directly from these features, to round out your text processing toolkit. One, like grep is insanely powerful once you learn its basics; the other is the opposite: conceptually very simple, yet crucial when processing gene sequences.
Modifying: sed
If grep is equivalent to the Find feature on many everyday applications, then sed is like Search and Replace, but on steroids. sed stands for stream editor—a name that makes more sense now that we’ve talked about text data as something that “flows” from one program to another via pipes.
sed’s main function is to take a line of text, then modify it according to some rule that you give it. As with grep, there are lots of rules that you can specify, but for this section we will only highlight the ones that you’ll really need for the week’s assignments.
This is how a sed invocation looks:
sed "<rule>" <filename>
As you have now learned, you can also drop filename, making sed apply the rule to lines of text that you type directly, one line at a time:
sed "<rule>"
This latter form of sed can also be used as part of a pipe:
grep "<pattern>" <filename> | sed "<rule>"
The above pipe will extract the lines in filename that match pattern, then make sed modify those lines according to the given rule.
- Note: For now, the way we are using sed does not make any permanent changes to files, so don't worry about messing up any of the data that you’re using. All examples just display the edited text. There is a way to make sed’s modifications permanent, but that’s beyond the scope of what we want to do for now.
Replacing Text That Fits a Pattern
Perhaps the most commonly-used modification rule in sed is s/<pattern>/<replacement>/g. The use of the term pattern is no accident—the patterns that sed recognizes for matching text are nearly identical to those recognized by grep. replacement then takes the place of those matched patterns of text.
Thus, a full text replacement invocation of sed, without a filename, looks like this:
sed "s/<pattern>/<replacement>/g"
Give this a shot:
sed "s/Hello/Goodbye/g"
If you then type Hello, hi, bye, and Hello World! as individual lines, sed interjects its output to produce this:
Hello Goodbye hi hi bye bye Hello World! Goodbye World!
Observe that, unlike grep, sed does repeat every line you type, regardless of whether or not that line matches the pattern. The difference is that, when there is a match, sed performs the specified replacement. Thus, Hello becomes Goodbye, and Hello World! becomes Goodbye World!
The power behind this search-and-replace functionality comes from the patterns that you can use, again very similar to those used by grep. In addition to exact matches (like the one showed above), you can do:
. Matches a single character [<characters>] Matches lines that have any of the characters listed in <characters> ^pattern Matches lines that start with the given pattern pattern$ Matches lines that end with the given pattern pattern* Matches zero or more repetitions of the given pattern
Instead of absolute negation (-v in grep), sed attaches a different meaning to ^ when it’s included in between brackets [ ]: sed "s/[^FLIMVSPTAYHQNKDECWRG-]/*/g" ...replaces any letter that is not one of the characters in between the brackets with an asterisk (*).
In terms of replacement, in addition to replacement with an exact piece of text, you can also delete the matched text; this is a matter of placing nothing in between the second set of slashes (/):
sed "s/Evil//g"
...deletes the text “Evil” from any input lines that have it. Of course, you can use non-exact patterns, such as this:
sed "s/^..//g"
The above rule unconditionally deletes the first two characters of each input line, no matter what that character is.
Perhaps even more powerful, you can also include the matched text—even though you don’t know what that is per line—in the replacement. Do this by including an ampersand (&) in the replacement text. The matched text replaces the ampersand in the final output. For example:
sed "s/.../& /g"
...this will replace any three characters with the same characters plus a space. Since those three characters will differ from line to line (and in fact many lines will have more than one set of three characters), having & available lets you keep those while adding a space.
As another example:
sed "s/[aeiou]/&&/g"
...will double up every lowercase vowel in the input text.
Try those search-and-replace operations on Microsoft Word :) Not impossible, but probably harder than doing it with sed (once you learn the patterns).
As mentioned, there’s definitely a lot more that you can do, but this should be enough to get you through the immediate assignments.
Gathering Up a Bunch of Rules in a Single File
What if you want to perform a whole bunch of search/replace activities on some text data? On the one hand, you can type multiple sed commands in a pipe. For example, changing all “The”s to “Them” then changing “Bones” to “Brains” may be done this way:
sed "s/The /Them /g" | sed "s/Bones/Brains/g"
When you have a lot of substitutions to do—like, say, replacing mRNA base triplets with their corresponding amino acid letters—it would be a pain to write out a long pipe (64 sed commands, to be exact, in the case of mRNA-to-amino acid translation).
For precisely this reason, sed allows a variation that does not include the actual rule in the command, but reads the rules from a separate file:
sed -f <file with rules>
The file with rules is a simple text file, with one sed substitution rule per line. Invoking sed -f <file with rules> on a stream of text data is equivalent to performing sed, sequentially, once for every rule in file with rules. It’s mainly a time saver, but a significant one.
Creating this file is another story, and is beyond the scope of this page. The one case in your assignments where sed -f <file with rules> will be useful is precisely when transcribing the genetic code. For this reason, a sed rules file, listing these substitutions for lowercase bases converting into uppercase amino acids, has been pre-created for you. You’ve seen this file before: it’s ~dondi/xmlpipedb/data/genetic-code.sed.
At this point, your working directory should be ~dondi/xmlpipedb/data. Thus, just typing this should allow you to look those rules over, and verify that they indeed represent the genetic code:
more genetic-code.sed
If pwd does not display /nfs/home/dondi/xmlpipedb/data, then do this first, and then invoke the more command above:
cd ~dondi/xmlpipedb/data
Replacing Characters With Another Set of Characters
As powerful as s/<pattern>/<replacement>/g it is, it actually has limitations. For example, what if you wanted to do something similar to a “secret decoder ring,” where, say, every letter becomes the letter after it, and “z” cycles back to “a”? You might think that including a sequence of s/<pattern>/<replacement>/g rules in a file will do this, but it won’t (for simplicity, we’re only including lowercase):
s/a/b/g s/b/c/g s/c/d/g ... s/z/a/g
This won’t work: since the replacements are done in sequence, a word like “adios” then becomes “bdios” after the first substitution (i.e., “b” for “a”). Then, when “b” is substituted for “c”, “bdios” then becomes “cdios”—which isn’t what you want.
What we need is a different rule, which substitutes multiple letters for a different one in one fell swoop. This rule does exist in sed, and that is:
y/<original characters>/<new characters>/
Because the replacement must be one-to-one, there must be as many characters in <original characters> as there are in <new characters>. With the y/<original characters>/<new characters>/ rule, the “secret decoder ring” becomes possible:
sed "y/abcdefghijklmnopqrstuvwxyz/bcdefghijklmnopqrstuvwxyza/"
As you might expect, this sed command will “decode” the message produced by the one above:
sed "y/bcdefghijklmnopqrstuvwxyza/abcdefghijklmnopqrstuvwxyz/"
Inclusion of uppercase letters, plus any other substitutions, are left to you for practice. Do note, however, how y/<original characters>/<new characters>/ is materially different from s/<pattern>/<replacement>/g.
Reversing: rev
At this point, the power and flexibility of grep, piping, and sed can be rather overwhelming. Thus, we’ve saved the last command for something relatively simple—text reversal. The rev command is refreshingly simple, with the only variation being whether or not it reverses a pre-existing text file or something that you type directly:
rev <optional filename>
If you type this (while having ~dondi/xmlpipedb/data as your working directory):
rev genetic-code.sed
...you’ll see each line of that file reversed. Of course, sed won’t be able to use this file anymore, but the point for now is to show what rev does.
Alternatively, typing rev all by itself leads to the now-familiar direct text entry mode:
rev
...whereupon typing the text “Oh say can you see” and “by the dawn's early light” results in:
Oh say can you see ees uoy nac yas hO by the dawn's early light thgil ylrae s'nwad eht yb
Of course, rev can fully participate in a pipe. This command sequence extracts movies with the word “Vampire” from the movie_titles.txt file, removes their IDs and release years, then reverses the resulting titles:
grep "Vampire" movie_titles.txt | sed "s/^.*,....,//g" | rev
Simple, but useful—especially when working with gene sequences.
Concluding Summary
This is a lot to digest at once, so here are the main points to take home:
- Command line interaction is a cycle of typing in commands then viewing their results
- Assorted shortcuts, like a history and the Tab key, try to minimize the pain of constant typing
- Files and folders/directories are available on the command line just like on any other computer, but are just expressed and accessed without graphics
- Files and their locations are represented as paths
- At any time, there is one working directory
- pwd displays the working directory, while cd changes it
- Text processing commands provide powerful ways for viewing, finding, and changing text
- cat displays text continuously; more breaks it up into screenfuls
- wc displays the number of lines, words, and characters in a piece of text data
- grep filters out lines of text according to a pattern
- sed replaces various patterns of text with some other text
- rev reverses lines of text
- Text processing commands can be “strung together” in a pipe, thus enabling even more text operations
- The vertical bar character ( | ) is used to indicate that one command should “pipe” its output as input to the next one
Examples
Will all of this in mind, here is a list of example commands, with a description of what they do. To promote experimentation, these examples all expect directly-typed text as input; to use another file, include a filename as part of the first command in the sequence.
- This command counts the number of lines that include the word “Count” (the number of words in these lines, plus their total number of characters, is also included)
grep "Count" | wc
- This command replaces the word “money” with “bucks”
sed "s/money/bucks/g"
- This command replaces the word “money” in any mixture of upper- or lowercase with “bucks”
sed "s/[Mm][Oo][Nn][Ee][Yy]/bucks/g"
- This command performs “Rot-13” encryption on lowercase characters: the same command is used for both encoding and decoding a message (can you see why?)
sed "y/abcdefghijklmnopqrstuvwxyz/nopqrstuvwxyzabcdefghijklm/"
- This command chops off the last word of every line
sed "s/ [^ ]*$//g"
- This command extracts lines that start with the uppercase letter “A”, adds a “0” after every fourth character, then reverses the line
grep "^A" | sed "s/..../&0/g" | rev
- This command capitalizes all “a”s and “t”s in the text, then removes them
sed "y/at/AT/" | sed "s/[AT]//g"
Additional Material
Interested in deepening your command line fu? Additional handouts can be found here: