Emilysimso Week 9
Contents
- 1 Directions
- 1.1 Download and Extract Data Source Files
- 1.2 Download or Update GenMAPP Builder
- 1.3 Create New Database in PostgreSQL
- 1.4 Configuring GenMAPP Builder to Connect to your PostgreSQL Database
- 1.5 Importing Data into the PostgreSQL Database
- 1.6 Tally Engine
- 1.7 XMLPipeDB Match
- 1.8 SQL
- 1.9 Microsoft Access
- 1.10 Back to the Command Line
- 2 Notes
- 3 OriginalRowCounts Comparison
- 4 Visual Inspection
- 5 .gdb Use in GenMAPP
- 6 Compare Gene Database to Outside Resource
- 7 Weekly Assignment Information
Directions
Download and Extract Data Source Files
- Downloaded UniProt XML, GOA, and GO OBO-XML files.
UniProt XML
- Went to the UniProt Complete Proteomes page.
- Browsed to get to Vibrio cholerae page, first filtered the list by clicking on the link for "Bacteria" under the "Superkingdom" heading.
- Further filtered the results for those species with a "Reference proteome".
- Scrolled through the results until I found Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961).
- Clicked on the link for "UniProtKB", e.g. Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961).
- Clicked the "Download" button at the top of the page and selected the following options:
- Selected the radio button to "Download all"
- Chose "XML" from the "Format" drop-down menu.
- Selected the radio button for "Compressed" format.
- Clicked the "Go" button.
GOA
- This is the UniProt-GOA home page.
- The current and previous UniProt-GOA files can be downloaded from the UniProt-GOA ftp site.
- In the directory that appears, clicked the link to the "proteomes" directory.
- Note that it may take some time to load this page.
- Found my organism of interest and right-clicked on the link to download the GO annotations and selected "Save target as" or "Save link as" and saved the GOA file. For example, this is the link for Vibrio cholerae.
- Note: Since the GOA file is a text file, your browser will not automatically download it when you left-click on the link. Instead, it will try to open the file in your browser window. Since it is a large file, this could take a long time if your internet connection is slow.
- The version information can be found on displayed in the ftp file directory under the "Last modified" column (Needed for my Gene Database Testing Report)
GO OBO-XML
- Downloaded the GO OBO-XML formatted file from the Gene Ontology download page. Clicked on the link for "obo-xml.gz" under the heading "Legacy Downloads."
- This file is updated daily. You can get the day/time that the file was created from the file properties after you have unzipped the file.
Extract the UniProt XML and GO OBO-XML files
- Extracted the UniProt XML and GO OBO-XML .gz files using 7-zip
Download or Update GenMAPP Builder
- Visited the XMLPipeDB releases page on GitHub.
- Extracted the GenMAPP Builder folder using 7-zip or other utility.
- We suggest that you move the extracted folder to the "T:" drive on the computers in Seaver 120. This folder is a "Thawspace", the contents of which will not be deleted by the program Deep Freeze when the computer is restarted.
Create New Database in PostgreSQL
NOTE: if you have already performed this step and want to use GenMAPP Builder functions with a database you previously created in PostgreSQL, you can skip this step.
- Launched pgAdmin III.
- Double-clicked on PostgreSQL 9.4 (localhost:5432) on the upper left hand side of the window.
- Right clicked on "Databases" and Selected "New Database..."
- Gave the database a name in the "Name" field and clicked OK (named Esimso V. cholerae 20151027 GVMBbuild5)
- Double-left-clicked on my new database name in the treeview on the left.
- Clicked on the SQL icon in the toolbar at the top of the window.
- The SQL Editor tab opened and there was leftover query text in the upper pane. Deleted this text. You are now going to use an XMLPipeDB query to create the tables in the database.
- Clicked on the Open File icon in the toolbar (the yellow folder with an arrow).
- Navigated to the folder in which I unzipped GenMAPP Builder.
- Opened the sql folder and opened the file gmbuilder.sql.
- Clicked the Execute Query icon, which looks like a green "Play" triangle button.
- This query now created all the tables in the database (although there is still no data in them).
- Closed the query window
- To double check that all was OK, clicked the + sign for the database, then the + sign for Schemas, then finally the + sign for public. Under the Tables section, I saw a count of 167 in parentheses.
Configuring GenMAPP Builder to Connect to your PostgreSQL Database
- Launched gmbuilder.bat.
- Selected the menu item File > Configure Database...
- Under the Database Connections tab the Database Driver defaults to PostgreSQL. Entered information in the following fields:
- Host or address: localhost
- Port number: 5432
- Database name: <enter the name of the PostgreSQL database you created above>
- Username: <enter the username of the PostgreSQL database you created above>; in S120, this username is "postgres"
- Password: <enter the password of the PostgreSQL database you created above>; in S120, ask the instructors for the password.
- Clicked the OK button.
Importing Data into the PostgreSQL Database
- Selected File > Import UniProt XML...
- Navigated to the UniProt XML file that I extracted previously and clicked the Open button.
- This should take about 5-10 minutes, but may take longer depending on the size of the file, processor speed, and available memory of the machine. When the process completed, recorded the elapsed time from the message window that appears.
- Selected File > Import GO OBO-XML...
- Navigated to the GO OBO-XML file that I extracted previously. Clicked the Open button.
- This should take about 5-10 minutes, but may take longer depending on the size of the file, processor speed, and available memory of the machine. When the process completed, recorded the elapsed time from the message window that appears.
- Clicked OK to the message asking me to process the GO data.
- This should take about 5-10 minutes, but may take longer depending on the size of the file, processor speed, and available memory of the machine. When the process completed, recorded the elapsed time from the message window that appears.
- Selected File > Import GOA...
- Navigated to the GOA file that I downloaded previously and clicked the Import button. This process only took a minute or so.
Tally Engine
The first tool, called the Tally Engine, can be used for verifying that certain data from the XML file transferred consistently into the PostgreSQL database upon import. The Tally Engine can be found in GenMAPP Builder itself.
- Ran PostgreSQL (via pgAdmin III on Windows) and make sure that your database is up and running.
- Ran GenMAPP Builder and make sure that it is connected to the database (via Configure Database...).
- After performing an import, chose Run XML and Database Tallies for UniProt and GO....
- Chose the UniProt and GO files that I imported.
Under the hood, the Tally Engine bases its XML counts on certain XML tags, and bases its database counts on SQL queries using count. This tool is thus primarily useful for making sure that the “raw” import worked without any errors or glitches.
XMLPipeDB Match
XMLPipeDB Match is useful for counting data in files. Thus, in our context, you would use XMLPipeDB Match to tally stuff in XML files, with greater flexibility than with Tally Engine.
You will have to use XMLPipeDB Match from the command line. In addition, you can use this in any platform (as you have seen). Download the application from the XMLPipeDB SourceForge site and take note of the location of the xmlpipedb-match-1.1.1.jar. Then, on the command line (Terminal for Linux and Mac OS X, cmd on Windows), cd to the folder containing the XML file that you would like to check. Use XMLPipeDB Match as follows, with the parts in parentheses varying depending on your specific setup, desired pattern, and file being scanned:
java -jar (location-of-jar) "(pattern)" < (XML file)
On a Windows machine, with XMLPipeDB Match and a Vibrio cholerae XML file located on the Desktop, scanning for IDs of the form VC_####, where # represents a digit from 0 to 9, one would type, after cd-ing to the Desktop:
java -jar xmlpipedb-match-1.1.1.jar "VC_[0-9][0-9][0-9][0-9]" < uniprot-taxonomy%3A243277.xml
As you have seen before, this will give you a list of unique matches, with a total number at the bottom.
The trick with XMLPipeDB Match is to use the patterns well: with the database project, you will mainly be matching IDs. A desired count is an XMLPipeDB Match result whose matched ID pattern corresponds to the number of IDs found by the Tally Engine.
SQL
You can also look for counts at the SQL level, using some variation of a select count(*) query. This requires some knowledge of which table received what data. Here’s an initial tip: the gene/name tags in the XML file land in the genenametype table. A query on this table counting values from this table that were marked as ordered locus in the XML file matching the pattern VC_[0-9][0-9][0-9][0-9] would look like this:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';
In pgAdmin III, you can issue these queries by clicking on the pencil/SQL icon in the toolbar, typing the query into the SQL Editor tab, then clicking on the green triangular Play button to run.
Microsoft Access
For the GenMAPP Gene Database, you can open the .gdb in Microsoft Access and navigate its tables to find counts for various IDs. Opening the table, noting its size, and doing some sorting may help. You can also look at the OriginalRowCounts table for a summary of totals.
Again, the ideal situation is a correspondence in these numbers with what you found in XML and the relational database.
Back to the Command Line
Amidst all this, you can still use grep and wc on the command line for some basic counting. Just remember that these tools work on a line-by-line basis; useful in some cases, but not useful in others.
You can use grep and wc with the various files on the my.cs.lmu.edu server by using the curl -O command shown in the Week 6 assignment. Upload your data files to the wiki, place media links to them on your wiki page, then mouse over those live links to capture their URL (like this—look at the source to see the wiki markup), then use curl -O (whatever-the-url-is) while ssh-ed to my.cs.lmu.edu to bring that file into the server.
- If the file is a .zip file, you can use unzip at the command line to unzip it.
- If the file is a .gz file, you can use gunzip at the command line to uncompress that one.
Notes
Version of GenMAPP Builder: gmb3build5
Computer on which export was run: HP LV2311
Postgres Database name: Esimso V. cholerae 20151027 GVMBbuild5
UniProt XML filename - uniprot-organism%3A243277.xml NEED LINK:
- UniProt XML version (The version information can be found at the UniProt News Page):
- UniProt XML download link:
- Time taken to import: 3.14 minutes
- Note: the Wiki was unable to upload the file
GO OBO-XML filename - go_daily-termdb.obo-xml NEED LINK:
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped):
- GO OBO-XML download link:
- Time taken to import: 7.36 minutes
- Time taken to process: 4.37 minutes
- Note: took a long time to import
- Note: the Wiki was unable to upload this file as well, as it was in the incorrect form
GOA filename - 46.V_cholerae_ATCC_39315.goa File:46.V cholerae ATCC 39315 ES.goa.txt:
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site):
- GOA download link:
- Time taken to import: 0.07 minutes
- Note:
Name of .gdb file - Vc-Std_20151027_ES.gdb File:Vc-Std 20151027 ES.gdb:
- Time taken to export:
- Start time: 3:52:08 PM
- End time: 5:14:19 PM
- Note: had to leave due to class ending so checked back next class period
TallyEngine
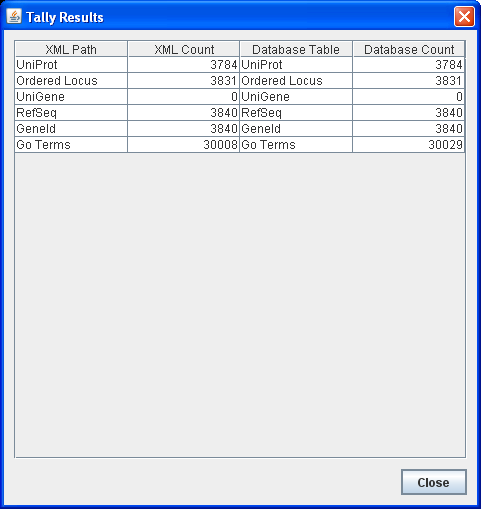
- Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases.
- Choose the menu item Tallies > Run XML and Database Tallies for UniProt and GO...
- Take a screenshot of the results. Upload the image to the wiki and display it on this page.

- For more information, see this page.
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Follow the instructions found on this page to run XMLPipeDB match.
Are your results the same as you got for the TallyEngine? Why or why not?
- XML had 2738 unique matches, which does not match from TallyEngine (had 3831 Ordered Locus hits). These may be different if there were multiple entries that were the same but that XML counts as one.
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
For more information, see this page.
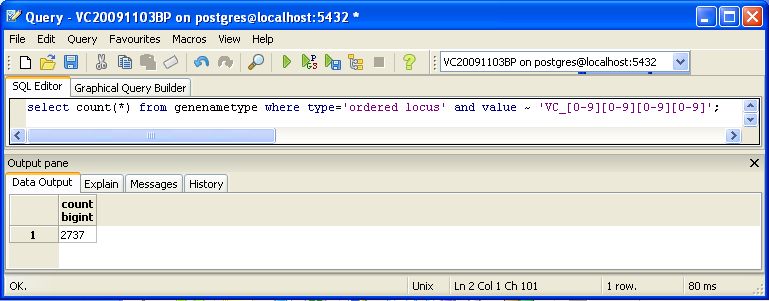
You can also look for counts at the SQL level, using some variation of a select count(*) query. This requires some knowledge of which table received what data. Here’s an initial tip: the gene/name tags in the XML file land in the genenametype table. A query on this table counting values from this table that were marked as ordered locus in the XML file matching the pattern VC_[0-9][0-9][0-9][0-9] would look like this:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';
In pgAdmin III, you can issue these queries by clicking on the pencil/SQL icon in the toolbar, typing the query into the SQL Editor tab, then clicking on the green triangular Play button to run.
Are your results the same as reported by the TallyEngine? Why or why not?
- Got 2727 results from SQL search. Again, this is not the same as TallyEngine (had 3831).
- Microsoft Access had 7664 OrderedLocusNames results from gdb.
- Some IDs start with VC, others with VCA, some have VC_ and others VC
- 7664 is roughly double 3831 - there may be multiple versions of the same table or entry
- Tested this and saw that there were 4 entries for one ID (tested 0703)
- Microarray data does not have an underscore
- Differences between human and computer understanding, had to populate with both versions of the data
- Used [select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]' or value ~ 'VC_A[0-9][0-9][0-9][0-9]';] in SQL
- Got 3831 results
- Also tried [select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9]';]
- Got 3831 results
- Ran same thing in "match" and got 3831 as well
- So VC_A?[0-9][0-9][0-9][0-9] is the true pattern
- But there are two additional entries in Access (7664)
- Tested using Xcel
- Note: transfer from match using >
- Problems occurred at VC_A0360.1
- Tested using Xcel
OriginalRowCounts Comparison
Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
Benchmark .gdb file:
Copy the OriginalRowCounts table from the benchmark and new gdb and paste them here:
- This screenshot shows the first portion of the rows from the gdb file

- This screenshot shows the screenshot from the benchmark NEED FILE
Note:
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
- There are not dates present for all of the gene ID systems
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
- There appears to be some discrepancies between the IDs present. For example, in the OrderedLocusNames table, some of the IDs contain an A while others do not, or some have a "_" that is not present in all IDs.
Note:
.gdb Use in GenMAPP
While the above sections perform quality assurance on the exported Gene Database via verifying ID counts, the "proof in the pudding" is to actually use the Gene Database in GenMAPP. You can follow the instructions in Part 2 of the Vibrio cholerae Microarray Data Analysis to verify that the Gene Database works in GenMAPP. In this case, the emphasis is not on the findings of the data analysis itself, but that the Gene Database functions appropriate in GenMAPP.
Note:
Putting a gene on the MAPP using the GeneFinder window
- Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
Note: there were 3831 errors loading the IDs into MAPP
Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
- 3831 IDs were uploaded (used the .txt file and removed the headers from the columns)
- I do not think there were any exceptions
- The error code is that "Gene ID system 2 not in Gene Database"
- I do not believe were not present in the UniProt XML
Note:
Coloring a MAPP with expression data
Note:
Running MAPPFinder
Note:
Compare Gene Database to Outside Resource
Note: This section applies to the Group Final Project and does not need to be completed for the Week 9 assignment. — Kdahlquist (talk) 15:46, 2 November 2015 (PST)
The OrderedLocusNames IDs in the exported Gene Database are derived from the UniProt XML. It is a good idea to check your list of OrderedLocusNames IDs to see how complete it is using the original source of the data (the sequencing organization, the MOD, etc.) Because UniProt is a protein database, it does not reference any non-protein genome features such as genes that code for functional RNAs, centromeres, telomeres, etc.
Note:
Weekly Assignment Information
Assignments
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 13
- Week 14
- Week 15
Individual Journal Entries
- Emilysimso Week 2
- Emilysimso Week 3
- Emilysimso Week 4
- Emilysimso Week 5
- Emilysimso Week 6
- Emilysimso Week 7
- Emilysimso Week 8
- Emilysimso Week 9
- Emilysimso Week 10
- Emilysimso Week 11
- Emilysimso Week 12
- Emilysimso Week 13
- Emilysimso Week 14
- Emilysimso Week 15
Class Journal Entries
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Class Journal Week 10
- Class Journal Week 11
- Class Journal Week 12
- Class Journal Week 13
- Class Journal Week 14
- Class Journal Week 15