Ksherbina Project Notebook
| Assignment Description | Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 | Week 11 | Week 12 | Week 13 | Week 15 |
| Class Journal | Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | |||||
| Individual Journal | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 | Week 11 |
| Other | Week 5: Database Wiki |
| Final Project | Team H(oo)KD Project Page | Journal Club Presentation | Project Individual Journal |
Contents |
Week 12
Starting the Set Up of the Testing Environment
- Downloaded the following software onto my personal computer:
- Latest version of GenMAPP Builder (gmbuilder2.0-b71) from SourceForge
- Java SE 7u45
- Eclipse IDE for Java EE Developers
- Created an account on SourceForge.
Import/Export of Gene Database
- Downloaded the UniProt XML for C. trachomatis serovar A strain HAR-13: Uniprot XML C.trachomatis serovar A KS 20131114.xml
- Downloaded the GOA file for C. trachomatis serovar A: 22183.C trachomatis A KS 20131114.goa
- Created a new database in pgAdminIII: CT_KS_20131114_gmb2b71
- Created tables in the database by executing the script in the file gmbuilder.sql.
- Launched gmbuilder-32bit.bat:
- Configured the database in gmbuilder:
- Host or address: localhost
- Port number: 5432
- Database name: CT_KS_20131114_gmb2b71
- Username: postgres
- Password: <password of the PostgreSQL database created above>
- Imported the UniProt XML file into the PostgreSQL database through gmbuilder.
- Imported the OBO-XML file into the PostgreSQL database through gmbuilder.
- gmbuilder was not able to completely process the data. See the testing report below for details.
- Imported the GOA file into into the PostgreSQL database through gmbuilder.
- I did not begin the actual export of the database due to the problem with processing the OBO-XML file.
Testing Report
Version of GenMAPP Builder: 2.0b71
Computer on which export was run: Personal computer
Postgres Database name: CT_KS_20131114_gmb2b71
UniProt XML filename: Uniprot_XML_C.trachomatis_serovar_A_KS_20131114.xml
- UniProt XML version (The version information can be found at the UniProt News Page): UniProt release 2013_11
- Time taken to import: 1.70 min
GO OBO-XML filename: Go_daily-termdb_v2_HD_20131107.obo-xml
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped):
- Time taken to import: 9.29 min
- Time taken to process: Could no complete the process. The following error came up when gmbuilder was processing the 81000 term:
ERROR edu.lmu.xmlpipedb.gmbuilder.GenMAPPBuilder - java.util.concurrent.ExecutionException:java.lang.OutOfMemoryError: Java heap space
GOA filename: 22183.C_trachomatis_A_KS_20131114.goa
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site):
- Time taken to import: 0.02 min
Name of .gdb file:
- Time taken to export .gdb:
- Upload your file and link to it here.
Note:
Creating Custom Species Profile
- On one of the class computers in the first row, I launched Eclipse with Subclipse.
- Went to Window > Open Perspective > Other > SVN Repository Exploring.
- Defined a new subversion by clicking on Add Repository and adding the URL https://svn.code.sf.net/p/xmlpipedb/code. Clicked Finish.
- Double-clicked on trunk, right-clicked on gmbuilder, and chose Checkout....
- Created a new project (chose Check out as a project configured using the New Project Wizard).
- Went to Window > Preferences > Java > Installed JREs. Added a new environment with JDK.
- Double-clicked on the project folder.
- Double-clicked on the lib folder.
- Shift-clicked and selected all files in the lib folder and then control-clicked on every file inside the lib folder whose name does not end in .jar. Right-click on one of them and choose Build Path > Add to Build Path from the popup menu that comes up.
- Created a species profile called ChlamydiaTrachomatisSerovarA.
- Dondi modified the name to ChlamydiaTrachomatisSerovarAUniProtSpeciesProfile.
- Customized the species profile.
- Opened the new profile and added the lines of code as specified in "Customize the Species Profile" directions.
- Still need to insert the species-specific URL that returns a web page describing a gene for that species.
- Opened the new profile and added the lines of code as specified in "Customize the Species Profile" directions.
- Committed the profile following the directions in "Updating and Committing Code".
Reflection
- What were the week’s key accomplishments?
- One of this week's key accomplishments was downloading the files necessary to run a import/export gene database cycle and the microarray files that will be analyzed. Also this week, we were able to start an import/export cycle.
- What are next week’s target accomplishments?
- Next week, I would like to complete an import/export cycle and vet it with the QA person. In addition, I hope that we can download the software necessary to open the microarray files, format the files, and perform statistical analysis.
- What team strengths were seen this week?
- I believe our strength this week as a team was meeting outside of class to create milestones. This meeting results in a detailed calendar of intermediate milestones that we plan to meet accomplish in order to meet the file deadline.
- What team weaknesses were seen this week?
- It was difficult to set up the aforementioned meeting due to our busy schedules. Additionally, we did not have a clear plan for working on the project until the middle/end of the week. In the coming weeks, our goal will be to improve our work plan for each class period so as to minimize the difficulty of scheduling work meetings outside of class.
Week 13
November 15, 2013
Import/Export Gene Database
After the error that came up last week regarding the Java heap space, I performed the following steps to increase the heap space:
- Open up gmbuilder-32bit.bat in Notepad.
- Find the line
"C:\Program Files (x86)\Java\jre7\bin\java" -D32 -Xmx1024m -jar gmbuilder.jar
- Changed "-Xmx1024m" to "-Xmx2048m" thereby increasing the maximum heap space from 1024 MB to 2048 MB.
- Saved the file and closed it.
- Then, I tried the import/export again. I could not open gmbuilder-32bit.bat after I changed the maximum heap space. I repeated the above steps for gmbuilder.bat and found that I could then still open the program.
- Opening gmbuilder.bat, I imported both the UniProt XML file and the OBO-XML file. However, I received a Java heap space error again when processing the OBO-XML file.
- I increased the heap space in gmbuilder.bat from 2048 MB to 4096 MB and tried the import/export cycle again. When this did not work, I increased the heap space in gmbuilder.bat to 8192 MB.
- Then, I was able to import the UniProt XML, OBO-XML, and GOA files as well as process the OBO-XML.
- However, I was not able to begin the export because an error popped up stating that an export to a GenMAPP database is only possible in gmbuilder-32bit.bat.
Testing Report
Version of GenMAPP Builder: 2.0b71
Computer on which export was run: Personal computer
Postgres Database name: CT_KS_20131114_gmb2b71
UniProt XML filename: Uniprot_XML_C.trachomatis_serovar_A_KS_20131114.xml
- UniProt XML version (The version information can be found at the UniProt News Page):
- Time taken to import: 0.73 min
GO OBO-XML filename: Go_daily-termdb_v2_HD_20131107.obo-xml
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped):
- Time taken to import: 7.39 min
- Time taken to process: 47.15 min
GOA filename: 22183.C_trachomatis_A_KS_20131114.goa
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site):
- Time taken to import: 0.03 min
Name of .gdb file:
- Time taken to export .gdb:
- Upload your file and link to it here.
Note: Could not export the database. Received an error stating that an export to a GenMAPP database is only possible in gmbuilder-32bit.bat.
Troubleshooting
- Ran cmd from the Start menu.
- cd to where GenMAPP Builder is located.
- Typed the same command that is in the .bat file, but directly at the prompt:
"C:\Program Files (x86)\Java\jre7\bin\java" -D32 -Xmx1536m -jar gmbuilder.jar
Received the following error:
Error occurred during initialization of VM. Could not reserve enough space for project heap. Error: Could not create the Java Virtual Machine. Error: A fatal exception has occurred. Program will exit
November 19, 2013
Rerunning the Gene Database Import/Export
- From November 11/15/2013, the UniProt XML, OBO-XML, and GOA files were imported and the OBO-XML file was processed the in 64-bit gmbuilder.bat with the maximum heap space set to 8192.
- Performed the export in 32-bit gmbuilder.bat.
Testing Report
Version of GenMAPP Builder: Same as in previous testing report
Computer on which export was run: Same as in previous testing report
Postgres Database name: Same as in previous testing report
UniProt XML filename: Same as in previous testing report
- UniProt XML version (The version information can be found at the UniProt News Page):
- Time taken to import: 1.10 min
GO OBO-XML filename: Same as in previous testing report
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped):
- Time taken to import: 8.96 min
- Time taken to process: Did not reprocess because received a message stating that the GO data had already been processed.
GOA filename: Same as in previous testing report
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site):
- Time taken to import: 0.03 min
Name of .gdb file: Ct-Std v1 KS 20131119.gdb
- Time taken to export .gdb:
- Start Time: 4:01:10 AM
- End Time: 6:51:58 AM
- Upload your file and link to it here: Ct-Std v1 KS 20131119.gdb
Note:
Checking the Quality of the Exported Database
- In gmbuilder-32bit.bat, chose Run XML > Database Tallies for UniProt and GO....
- The XML count for the UniProt file was 917 while the Database Count for the Uniprot file was 10087. This number discrepancy is likely due to importing the UniProt, OBO-XML, and GOA files multiple times in experimenting with the heap space threshold.
- A new database must now be created in pgAdminIII in order to repeat the export/import gene database cycle.
November 20-21, 2013
Finishing Installing Software to Set Up the Development Environment
- Navigated to the "Download and Install" tab on the Subclipse home page
- Copied the Eclipse update site URL for the 1.10.x Release.
- Opened up Eclipse.
- Help > Install New Software...
- Pasted the URL into the "Work with" field.
- Checked the box next to "Subclipse" and "SVNKit".
- Clicked on Next and went through the process of installing the software.
Repeating the Gene Database Import/Export Cycle
- Created a new database in pgAdminIII.
- Created tables in the database by executing the script in the file gmbuilder.sql.
- Since the aforementioned error occurred because duplicates were introduced into the database through repeated imports, I decided to try this new import-export cycle using 32-bit gmbuilder.
- Opened gmbuilder-32bit.bat with the heap space set to 1024 MB and configured the new database.
- Host or address: localhost
- Port number: 5432
- Database name: CT_KS_20131119_32bit_gmb2b71
- Username: postgres
- Password: <password of the PostgreSQL database created above>
- Imported the UniProt XML file into PostgreSQL through gmbuilder-32bit.bat.
- Imported the OBO-XML file into PostgreSQL through gmbuilder-32bit.bat.
- Processed the OBO-XML file through gmbuilder-32bit.bat.
- Imported the GOA file into PostgreSQL through gmbuilder-32bit.bat.
Testing Report
Version of GenMAPP Builder: 2.0b71
Computer on which export was run: Personal computer
Postgres Database name: CT_KS_20131119_32bit_gmb2b71
UniProt XML filename: Uniprot_XML_C.trachomatis_serovar_A_KS_20131114.xml
- UniProt XML version (The version information can be found at the UniProt News Page): UniProt release 2013_11
- Original file name from UniProt site: uniprot-organism%3A315277+keyword%3A181.xml
- Time taken to import: 1.20 min
GO OBO-XML filename: Go_daily-termdb_v2_HD_20131107.obo-xml
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped): 11/06/2013
- Original file name: go_daily-termdb.obo-xml.gz
- Time taken to import: 13.05 min
- Time taken to process: 10.80 min
GOA filename: 22183.C_trachomatis_A_KS_20131114.goa
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site): 11/12/13
- Original file name: 22183.C_trachomatis_A.goa
- Time taken to import: 0.04 min
Name of .gdb file: Ct-Std KS 20131121.gdb
- Time taken to export .gdb:
- Start Time: 2:37:42 AM
- End Time: 2:55:45 AM
- Upload your file and link to it here: Ct-Std KS 20131121.gdb
Note: There were no heap space errors this time, which is good. The errors probably occurred in the first place because of repeatedly importing the files.
November 21, 2013
Revised the Custom Species Profile
- Ran Eclipse.
- Navigated to Window > Open Perspective > Other... and chose SVN Repository Exploring
- Defined a new Subversion repository by clicking on the Add Repository button.
- Set the URL to https://svn.code.sf.net/p/xmlpipedb/code then clicked Finish.
- Double-cliked on https://svn.code.sf.net/p/xmlpipedb/code in the list.
- Double-clicked on trunk.
- Right-clicked on gmbuilder and chose Checkout....
- Chose Check out as a project configured using the New Project Wizard then click Finish.
- In the New Project dialog that opened, chose Java Project from the list and then clicked Next >.
- Entered the new project name xmlpipedb-gmbuilder and clicked Finish.
- Set Eclipse to use JDK.
- Navigated to Windows > Preferences and clicked on the Search button.
- Expanded Computer > Local Disk (C:) > Program Files > Java > jdk1.7.0_45 and clicked OK.
- Highlighted the row jdk and clicked on Edit.
- In "JRE home:" field, removed everything in the name after jdk1.7.0_45 and then clicked Finish.
- Made sure that I was in the Java perspective by navigating to Window > Open Perspective > Other... and then choosing Java.
- Double-clicked on the xmlpipedb-gmbuilder folder.
- Double-clicked on the lib folder.
- Shift-clicked and selected all files in the lib folder and then control-clicked on every file inside the lib folder whose name did not end in .jar.
- Right-clicked on one of the files and choose Build Path > Add to Build Path.
- Check that the src folder was set to the source folder by right-clicking on it and then clicking on Build Path.
- Right-clicked on the test folder and chose Build Path > Use as Source Folder. Then, made sure that there were no red x's in the list of folders.
- Right-clicked on build.xml toward the bottom of the list. Then, navigated to Team > Synchronize with Repository.
- Double-clicked on src > edu.lmu.xmlpipedb.gmbuilder.databasetoolkit.profiles > ChlamydiaTrachomatisSerovarAUniProtSpeciesProfile.java.\
- Within the code, added the gene database link pattern: "http://bacteria.ensembl.org/chlamydia_trachomatis_a_har_13/Gene/Summary?g=~" and then saved the code.
- Right-clicked on build.xml and chose Run As > Ant Build... (the one with the ellipses).
- In the Edit Configuration dialog that appears, checked on the clean and dist items in the Targets tab.
- Clicked on the Order... button and rearranged the items so that clean is first and dist is second and then clicked OK.
- Clicked the Run button.
- Right-clicked on the xmlpipedb-gmbuilder project folder and chose Refresh (F5 is its keyboard shortcut).
Reran the Database Export With the New Build for gmbuilder
- Made sure that pgAdminIII and the database "CT_KS_20131119_32bit_gmb2b71" is open.
- Opened the newly built gmbuilder-32bit.bat within the folder [User Folder] > workspace > xmlpipedb-gmbuilder > dist.
- Made sure that the database was configured to "CT_KS_20131119_32bit_gmb2b71".
- Performed a database export.
Testing Report
- Name of .gdb file: Ct-Std_v2_KS_20131121.gdb
- Time taken to export .gdb:
- Start Time: 10:56 AM (approximately)
- End Time: 11:14:26 AM
- Upload your file and link to it here: Ct-Std v2 KS 20131121.gdb
Note:
Testing the New Database with GenMAPP
- Opened GenMAPP.
- Clicked on Data > New Gene Database and selected the gene database that was created today (Ct-Std v2 KS 20131121.gdb).
- On the toolbar, clicked on the button that has the word gene in a box.
- Clicked and dragged the cursor on the blank canvas to create a new gene box.
- Right-clicked on the gene box.
- In the pop-up window, typed in "CTA_0588" in the Gene ID search field.
- Under Gene ID System, selected Ordered Locus Names.
- Then, hit the "Search button". Interestingly, GenMAPP builder did not find this gene in the database.
- To troubleshoot, I opened up the gene database in Access.
- In the list of tables, I double-clicked on OrderedLocusNames.
- I sorted the list of gene IDs from A-Z and then scrolled down to try to find "CTA_0588". I was able to find "CTA_0587" and "CTA_0589" but not "CTA_0588".
- Back in GenMAPP, I used the same steps as above to find "CTA_0587".
- "CTA_0587" was found in the database. In the window, I clicked on the link "CTA_0587" under the OrderedLocusNames section.
- This link took me to the entry for this gene on the Model Organism Database (http://bacteria.ensembl.org/chlamydia_trachomatis_a_har_13/Gene/Summary?g=CTA_0587).
Performing Quality Assurance on the New Database
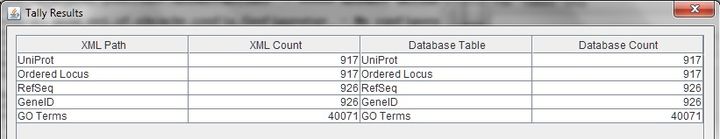
- Performed counts with Tally Engine.
- Counted the number of unique gene IDs using xmlpipdb match:
java -jar xmlpipedb-match-1.1.1.jar "CTA_[0-9][0-9][0-9][0-9]" < Uniprot_XML_C.trachomatis_serovar_A_KS_20131114.xml
- Total unique matches = 911
- Counted the number of unique gene IDs using an SQL query:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'CTA_[0-9][0-9][0-9][0-9]';
- Count - 917
- Opened the database in Access. Then, double-clicked on the table "OriginalRowCounts".
| Table | Rows |
| OrderedLocusNames | 919 |
| UniProt | 917 |
| UniProt-OrderedLocusNames | 919 |
- Double-clicked on the table "OrderedLocusNames" and scrolled all the way to the bottom. We found that some gene ID's had the format pCTA_####.
- Went back to the command prompt and ran the following command:
java -jar xmlpipedb-match-1.1.1.jar "[p|]CTA_[0-9][0-9][0-9][0-9]" < Uniprot_XML_C.trachomatis_serovar_A_KS_20131114.xml
- Total unique matches = 8
- Adding the results of the two runs with xmlpipedb match, we got 919 unique gene IDs, which is the same number as that in Access.
Week 14
November 24, 2013
Ran GenMAPP Using C. trachomatis Gene Database
- Opened GenMAPP.
- Made sure that the database was set to Ct-Std_v2_KS_20131121.gdb.
- Imported sample microarray data into GenMAPP.
- Loaded the microarray data by navigating to Expression Dataset Manager > New Dataset.
- There were 23015 errors.
- Opened the exceptions file that was created to inspect the errors and scrolled down to where the IDs CTA_#### begin.
- The gene IDs are actually in the format CTA_####_RRMH#####_at.
- Tried to search for a sample ID in the format found in the exceptions file in the UniProt database or any NCBI databases.
- Could not find CTA_0806_RRMH00303, RRMH00303, CTA_0806_RRMH00303_at, or RRMH00303_at in the UniProt database.
- Could not find CTA_0806_RRMH00303, RRMH00303, CTA_0806_RRMH00303_at, or RRMH00303_at in any database through NCBI.
- My suspicion is that the RRMH#####_at portion of the ID is an identifier that is appended to each probe on the Affymetrix microarray when it is read by the company's software.
November 27-28, 2013
Running SQL Queries to Determine Discrepancy in the Count Between the UniProt XML and the Gene Database
- Ran an SQL query to determine if there were any ordered locus names in the database that do not match the ID "CTA_####".
select * from genenametype where type = 'ordered locus' and not value ~ 'CTA_[0-9][0-9][0-9][0-9]';
- There were no hits.
- Ran an SQL query to show all the ordered locus names in the database that have the ID "CTA_####".
select * from genenametype where type = 'ordered locus' and value ~ 'CTA_[0-9][0-9][0-9][0-9]';
- Scrolling through the results, I saw the ID "CTA_0406/CTA_0407/CTA_0408".
- The UniProt manual stated that ordered locus names that are connected by a slash indicate two predicted genes that are actually one gene.
- The combination of these IDs explain why two less IDs show up in the count by the Tally Engine in comparison to the SQL query.
- Opened the gene database within Access and filtered the ordered locus names to show only those that match the ID "CTA_0407".
- The ID appeared on its own without being joined to other ID's with slashes.
Testing Report for Finalized Gene Database
Version of GenMAPP Builder: 2.0b71
Computer on which export was run: Personal computer
Postgres Database name: CT_KS_20131119_32bit_gmb2b71
UniProt XML filename: Uniprot_XML_C.trachomatis_serovar_A_KS_20131114.xml
- UniProt XML version (The version information can be found at the UniProt News Page): UniProt release 2013_11
- Original file name from UniProt site: uniprot-organism%3A315277+keyword%3A181.xml
- Time taken to import: 1.20 min
GO OBO-XML filename: Go_daily-termdb_v2_HD_20131107.obo-xml
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped): 11/06/2013
- Original file name: go_daily-termdb.obo-xml.gz
- Time taken to import: 13.05 min
- Time taken to process: 10.80 min
GOA filename: 22183.C_trachomatis_A_KS_20131114.goa
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site): 11/12/13
- Original file name: 22183.C_trachomatis_A.goa
- Time taken to import: 0.04 min
- Name of .gdb file: Ct-Std_v2_KS_20131121.gdb
- Time taken to export .gdb:
- Start Time: 10:56 AM (approximately)
- End Time: 11:14:26 AM
- Upload your file and link to it here: Ct-Std v2 KS 20131121.gdb
Note: There is one ID "CTA_0406/CTA_0407/CTA_0408" that is a combination of three predicted genes that are actually one gene. The Tally Engine counts this as one gene that contributes to the total count of 917. However, through Access and xmlpipedb match, the ID is separated into three separate genes bringing the total count to 919.
Week 15
December 3, 2013
- Worked with Hilda to separate the gene IDs from the Affymetrix IDs appended to them in the microarray data.
- Worked with Dillon to run GenMAPP and MAPPFinder with the microarray data and the gene database created for C. trachomatis.
December 5, 2013
Customize Tally Engine for C. trachomatis
- Opened Eclipse.
- Double-clicked xmlpipedb-gmbuilder > src > edu.lmu.xmlpipedb.gmbuilder.resource.properties > gmbuilder.properties.
- Found the following part of the code:
# # wizard.properties #
- Before this part of the code, added the following lines of code to specify which gene IDs to find in the UniProt XML file:
# Chlamydia trachomatis chlamydiatrachomatis_level_amount=1
chlamydiatrachomatis_element_level0=uniprot/entry/gene/name&type&ordered locus
chlamydiatrachomatis_query_level0=select count(*) from genenametype where type = 'ordered locus';
chlamydiatrachomatis_table_name_level0=Ordered Locus
- Saved the changes and built a new version of gmbuilder.
- Opened gmbuilder and set the database to CT_KS_20131119_32bit_gmb2b71.
- Ran TallyEngine.
- Got the same counts as those from November 21, 2013 despite the fact that I received an error that the species specified in the added code could not be found. However, the error also specified that the correct species ID is chlamydiatrachomatisservoara.
- Accordingly, I went back to Eclipse and modified the code added to gmbuilder.properties:
# Chlamydia trachomatis chlamydiatrachomatisserovara_level_amount=1
chlamydiatrachomatisserovara_element_level0=uniprot/entry/gene/name&type&ordered locus
chlamydiatrachomatisserovara_query_level0=select count(*) from genenametype where type = 'ordered locus';
chlamydiatrachomatisserovara_table_name_level0=Ordered Locus
- Built a distribution version of gmbuilder and then ran Tally Engine again. I got the same counts but no error message about the species ID as before.
- Went back to Eclipse. Synchronized the code on my computer with the code on SourceForge.
- Built a new distribution of gmbuilder.
- Synchronized again.
- Committed the changes I made to the gmbuilder.properties code.
Week 16
December 12, 2013
The gene database Ct-Std_v2_KS_20131121.gdb was renamed to Ct-Std_External_20131121.gdb in preparing the final delivarables for the project. The testing report was accordingly modified:
Testing Report
Version of GenMAPP Builder: 2.0b71
Computer on which export was run: Personal computer
Postgres Database name: CT_KS_20131119_32bit_gmb2b71
UniProt XML filename: Uniprot_XML_C.trachomatis_serovar_A_KS_20131114.xml
- UniProt XML version (The version information can be found at the UniProt News Page): UniProt release 2013_11
- Original file name from UniProt site: uniprot-organism%3A315277+keyword%3A181.xml
- Time taken to import: 1.20 min
GO OBO-XML filename: Go_daily-termdb_v2_HD_20131107.obo-xml
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped): 11/06/2013
- Original file name as listed in beta.geneontology.org: go_daily-termdb.obo-xml.gz
- Time taken to import: 13.05 min
- Time taken to process: 10.80 min
GOA filename: 22183.C_trachomatis_A_KS_20131114.goa
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site): 11/12/13
- Original file name as listed in the FTP site: 22183.C_trachomatis_A.goa
- Time taken to import: 0.04 min
- Name of .gdb file: Ct-Std_v2_KS_20131121.gdb
- Time taken to export .gdb:
- Start Time: 10:56 AM (approximately)
- End Time: 11:14:26 AM
- Upload your file and link to it here: Ct-Std_External_20131121.gdb
Note: There is one ID "CTA_0406/CTA_0407/CTA_0408" that is a combination of three predicted genes that are actually one gene. The Tally Engine and PostgreSQL count this as one gene resulting in a total gene count of 917. However, through Access and xmlpipedb match, the ID is separated into three separate genes bringing the total count to 919.