A Quick Relational Database Tour
This page gives you a tutorial-style walkthrough of a relational database, specifically PostgreSQL. The walkthrough assumes that you are at a Seaver 120 lab computer.
Contents
Overview
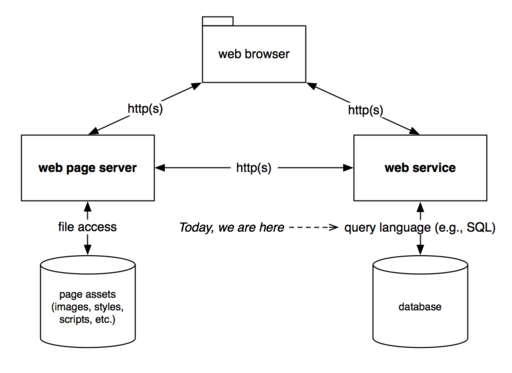
This diagram is a simplified rendition of how many web databases are structured. Most of the time, we only interact with these systems through a web browser. Later this semester, we will interact with the web service layer. This tutorial gives you a taste of what it’s like to interact directly with a database.
Running PostgreSQL on the Lab Machines
- Login to the computer as usual
- Launch pgAdmin
- The pgAdmin window starts with a hierarchical view on the left that starts with three layers:
- Browser
- Servers (1)
- PostgreSQL 9.6
- Servers (1)
- Browser
- Double-click on PostgreSQL 9.6 to connect to the database server
- The password to start the server will be given in class
Creating a Database
Once the server is running, the red x disappears from the PostgreSQL 9.6 icon, and additional icons appear beneath it. If you click on the + button to the left of the Databases icon, you will see the databases that are currently available. Initially, you will see a single database called postgres.
To create a database, right-click on the Databases icon and choose Create > Database... from the menu that appears. In the Create - Database dialog, the only information you need to supply is your new database’s name.
When you click Save, you will return to the main pgAdmin window and you should see your new database underneath the Databases icon.
Note that you only need to go through this creation process once; that database will remain available until it is explicitly deleted.
Connecting to a Database
To start using a database, click on its icon. The red x disappears from the database icon and you should now be able to work.
Creating a Table
It’s hard to use a database without any data—so let’s bring some in. For this tour, we’ll use some publicly-available data: the FDA Drug Database.
Download the linked .zip file in the Download File section of that page. Or curl it from the command line! You can use curl -O to automatically save the download as a file. To access the Windows Documents folder from bash, cd to /mnt/c/Users/biolab/Documents.
When you decompress the file, you’ll see that it’s a bunch of .txt files. Many databases are distributed in this format for maximum interoperability. However, this means that we have to do some work to get it into an actual relational database management system like PostgreSQL.
First, we have to know what is in the database. The FDA provides this in the “Data Definitions and ERD for Drugs@FDA” section of the above-linked page. If you click it, you’ll see a diagram of how the data in the files are related and the column definitions of each table.
We don’t have time to load everything, so we’ll just take one: the Products table. We see its definition on the page, and we can translate this into an SQL create table statement:
create table product(appl_no varchar(6) not null, product_no varchar(6) not null, form varchar(255), strength varchar(240), reference_drug int, drug_name varchar(125), active_ingredient varchar(255), reference_standard int, primary key (appl_no, product_no))
To do this in pgAdmin, choose Tools > Query Tool from the menu bar. A new Query tab appears. You can type (or copy-paste) the above command into that tab.
As always, watch out for typos! When ready, click on the Execute/Refresh button in the toolbar. (its button looks like a lightning bolt)
Upon executing the query, a success message should appear in the Messages tab of the Output Pane in the bottom half of the window.
To assure yourself that the product table is indeed there, type and execute this query:
select * from product
The Data Output tab of the Output Pane should now show an empty tabular display with headings for appl_no, product_no, form, strength, reference_drug, drug_name, active_ingredient, and reference_standard.
Inserting Data
The SQL insert statement allows you to add a record to a table. But if you look at the Products.txt file, you’ll notice that it is what’s called a tab-delimited text file. How does one build an insert statement for each of these lines without having to do a lot of typing? One solution involves a command you’ve seen before: sed!
Since this is a quick tour, we won't spend time to explain exactly how we come up with the sed command below. But you should be able to see the pieces:
- We need to turn all single apostrophes into doubles.
- We then need to “wrap” the tab-separated values at the end around single quotes and commas.
- We append the insert command before each line.
- We end each line with a parenthesis and semicolon.
cat Products.txt | sed "1d" | sed "s/'/''/g" | sed "s/\t/','/g" | sed "s/^/insert into product(appl_no, product_no, form, strength, reference_drug, drug_name, active_ingredient, reference_standard) values('/g" | sed "s/\r/');/g" | sed "s/'','');$/'');/g" | sed "s/'');$/'0');/g"
Try this command to see what comes out. One note of interest: you might have noticed that there are two extra sed steps in the command sequence above. These last two sed steps are there solely to accommodate some bad data in the file. You can try this whole sequence without those two steps to see what happens. Details in office hours if this intrigues you.
This command produces quite a few insert statements! To get them into PostgreSQL, you can save this into a file:
cat Products.txt | sed "1d" | sed "s/'/''/g" | sed "s/\t/','/g" | sed "s/^/insert into product(appl_no, product_no, form, strength, reference_drug, drug_name, active_ingredient, reference_standard) values('/g" | sed "s/\r/');/g" | sed "s/'','');$/'');/g" | sed "s/'');$/'0');/g" > Products.sql
Then, open the file in a text editor and copy/paste the commands into pgAdmin’s SQL Editor' tab. Click Execute query.
To save us some time, just like in a cooking show, we have a “baked version” of this file for your convenience. Download it from this link, unzip, then copy/paste into pgAdmin as stated above.
An Introduction to SQL Select Queries
Database activities are triggered via SQL commands. The select command is the SQL “kitchen sink” for retrieving information from a database. Its general, basic form is:
select columns from tables where conditions
As you will see, select can do even more, but let’s start simple.
Basics
The simplest type of select queries involve getting records from an individual table based on relatively simple conditions. For example:
select form from product where drug_name = 'ASPIRIN'
...will retrieve the method of delivery for drug products whose name is exactly “ASPIRIN.”
In addition to equality, like lends further flexibility. The like comparison allows for pattern matching, similar but not identical to grep. In SQL, the percent sign (“%”) is a “wildcard” that can represent any number of letters and symbols. like and % can be combined for broader queries, such as this one, which expands the previous query to include any drug whose name contains “ASPIRIN:”
select form from product where drug_name like '%ASPIRIN%'
If you want select to display all columns of a database record, use the asterisk (*):
select * from product where drug_name like '%ASPIRIN%'
Conditions can be combined via and and or; for example, retrieving drugs whose name contains either “ASPIRIN” or “TYLENOL” can be done with:
select * from product where drug_name like '%ASPIRIN%' or drug_name like '%TYLENOL%'
The like comparator can do simple text matches, but it does not use regular expressions (i.e., the search patterns recognized by grep and sed). This area is a little shaky in SQL-land; there is an official similar to comparator which is the official way to make regular expression comparisons, but the format for those expressions is not the same as the format used by grep and sed.
Fortunately, PostgreSQL has a specific, PostgreSQL-only comparator that does match the same patterns used by grep and sed: the tilde (~). Comparing with ~ is equivalent to a grep- or sed-like comparison:
select * from product where drug_name ~ '(DOPAMINE|NITRO)' select * from product where drug_name ~ 'ATE$' select * from product where strength ~ '^600'
The caveat here is that ~ is a PostgreSQL-specific feature: if you move to other database systems (such as Microsoft Access), that feature may either be done differently or missing completely, since it is not part of the official SQL standard.
Sorting
As you play with various queries on the product table, you’ve probably noticed that results are returned in no particular order; if you’d like to sort the results in some way, tack on an order by clause at the end of the select query:
select drug_name, form, strength from product where form like '%ORAL%' order by drug_name
You can add more fields for a very specific sort order:
select drug_name, form, strength from product where form like '%ORAL%' order by drug_name, form
The above query returns the same records, but this time sorted by drug_name first, then by form within the records with the same drug_name.
Sort order is ascending by default (e.g., A to Z, 0 to 9); for the reverse order, add desc to the field(s) that you’d like to see sorted in reverse:
select drug_name, form, strength from product where form like '%ORAL%' order by drug_name desc, form
This will display records with drug names displayed in reverse alphabetical order; within each drug name, however, form will still be sorted in ascending order.
Aggregate Queries
For large databases, SQL provides aggregate (a.k.a. “grouping”) queries that summarize multiple records in different ways.
The simplest form of summary is counting: how many records were retrieved? A simple overall count is done by using count(*) as the thing to select (or project, in formal relational algebraic terms):
select count(*) from product where drug_name like '%ASPIRIN%'
Of course, you can mix and match everything you have learned so far. To count the drug products whose names include “ASPIRIN” and are delivered by capsule, you can query:
select count(*) from product where drug_name like '%ASPIRIN%' and form like '%CAPSULE%'
Aggregators other than count are available, such as min, max, and avg (average or mean). We don’t have meaningful numeric fields in this table so we’ll leave that for another time.
Finally, you can aggregate multiple groups of data, so that you get different sets of statistics. The group by keyword does this:
select form, count(*) from product where drug_name ~ 'CODEINE' group by form
The main rule with group by is that the column being grouped should also be part of the select clause. This makes sense because otherwise, you wouldn’t be able to tell which group was which! And of course, you can mix and match. For example, the query above might yield more insight if we arrange the data by count:
select form, count(*) from product where drug_name ~ 'CODEINE' group by form order by count
Joins
Thus far, we’ve only been working with one table: product. The FDA drug database has 11 tables overall. The FDA drug database definition diagram shows how these tables relate to each other. SQL can join related tables together; although we do not have enough time to show this live (because it would entail loading additional tables plus having a deeper understanding of how to navigate those tables), the concept is important enough to at least describe.
An SQL join uses the same basic select command, but requires the tables being joined (such as product and application), as well as the fields/columns to use for “joining” records. “Joining” records means that their fields are combined to create a new “virtual” record. All other parts of the select command retain the same meanings as before:
select columns from table1 inner join table2 on (join condition) where conditions
As in the basic select command, you can tailor the where conditions as you need to pull records based on other criteria.
If you like what’s happening here, then solely for your personal edification, you can try to create and load all eleven tables from the FDA drug database into PostgreSQL. Slap a web service and some web pages on this database and you have yourself a data-driven web app!
The Notorious Apostrophe
You might have noticed that, because the apostrophe or single quote is used to indicate specific values in SQL (e.g., '%ASPIRIN%', 'CAPSULE', '^600', etc.), we run into a potential problem when the value itself should contain an apostrophe. This is not as uncommon as one might think; for example, a good number of drug products have apostrophes in their names (INFANTS' FEVERALL, CHILDREN'S MOTRIN, and WOMEN'S ROGAINE, to name a few). A statement such as the one below will not behave as you might expect, since the apostrophe will be misinterpreted as ending a piece of text rather than as part of the text itself:
select * from product where drug_name = 'CHILDREN'S ADVIL'
Fortunately, SQL has a solution: apostrophes inside text should be indicated via two consecutive apostrophes, or ''. When encountered, SQL converts this pair of apostrophes into a single one, and does not interpret these apostrophes as ending a piece of text. Thus, the above query will work if rewritten in this way:
select * from product where drug_name = 'CHILDREN''S ADVIL'
While the solution does exist, it isn’t automatic: you need to be aware that apostrophes have to be written as “double apostrophes” before passing any text values on to SQL. Keep this in mind when interacting with any database that uses SQL (which is to say, most relational database management systems).
SQL Injection
First, a little leadoff cartoon: http://xkcd.com/327
SQL injection is the term for a security exploit that attempts to bypass the web front end of a database and go directly to an SQL query. The exploit works on websites that do not—as stated in the XKCD strip—“sanitize their inputs.” In other words, a site is at risk for SQL injection if its query text field does not check for characters that might “break out of” the search term and thus spill into the SQL code.
This is probably best illustrated with an example…