HDelgadi Week 9
Export Information
Version of GenMAPP Builder: 2.0b70
- Computer on which export was run: First Computer second row on the left side.
Postgres Database name: vc_hdelgadi_20131022-gmb2b70
UniProt XML filename: File:Uniprot-organismHD 22102013.xml
- Followed UniProt XML on Running GenMAPP Builder
- UniProt XML version (The version information can be found at the UniProt News Page):
- Time taken to import: 8.90 minutes
GO OBO-XML filename: File:Go daily-termdb.obo-xml HD22102013.gz
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped):
- Time taken to import: 18.04 minutes
- Time taken to process: 14.08 minutes
GOA filename: File:46.V cholerae ATCC 39315 HD22102013.goa
- Direct download from wiki due to website error.
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site):
- Time taken to import: 0.20 minutes
Name of .gdb file: Vc-Std_20131022-HDelgadi-gmb2b70.gdb
- Time taken to export .gdb: Approximately one minute
- Upload your file and link to it here. File:Vc-Std 20131022-HDelgadi-gmb2b70.gdb
Note: Export started around 11 am; finished at 3:17:26 pm.
TallyEngine
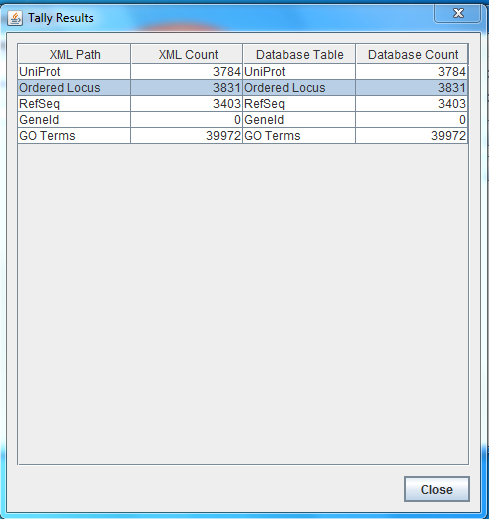
Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the PostgreSQL databases (or you can upload and link to a screenshot of the results).
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
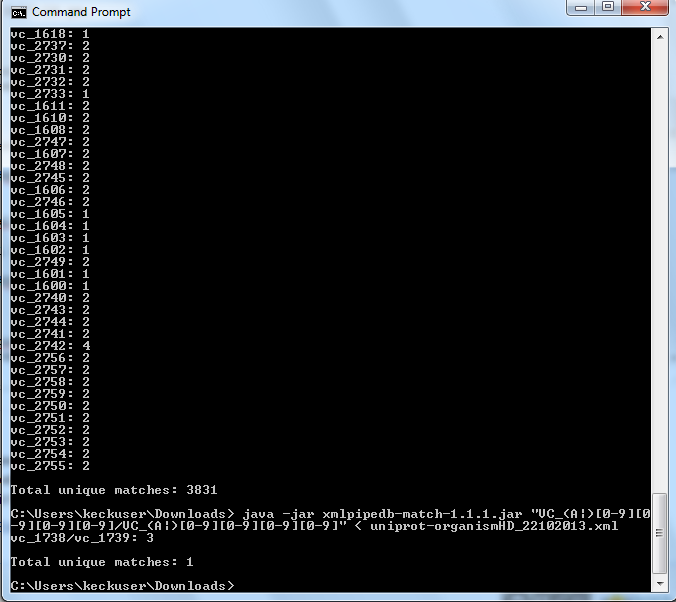
Follow the instructions found on this page to run XMLPipeDB match.
Are your results the same as you got for the TallyEngine? Why or why not?
The results are the same as the TallyEngine.
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
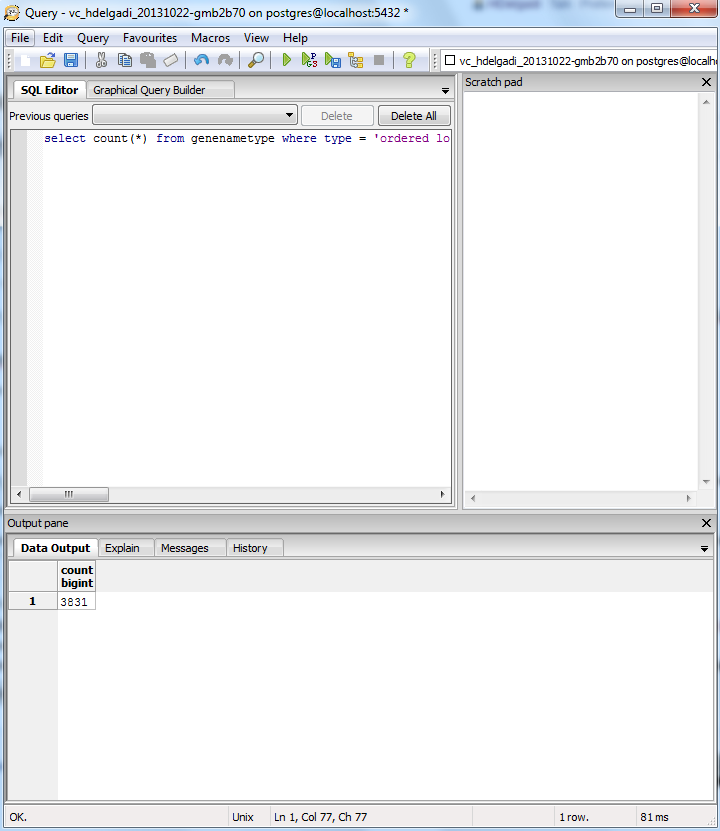
Follow the instructions on this page to query the PostgreSQL Database.
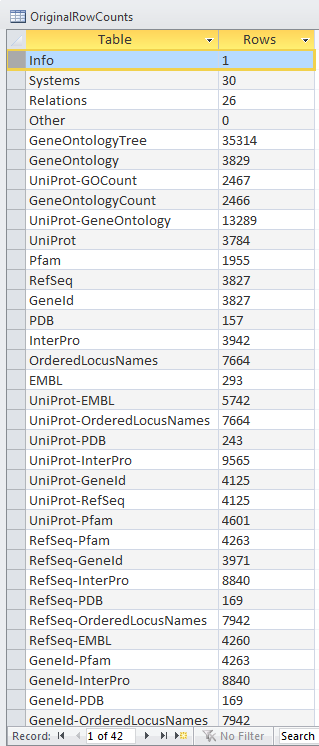
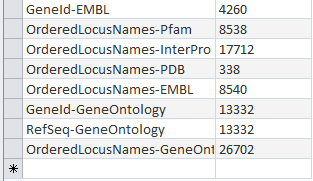
OriginalRowCounts Comparison
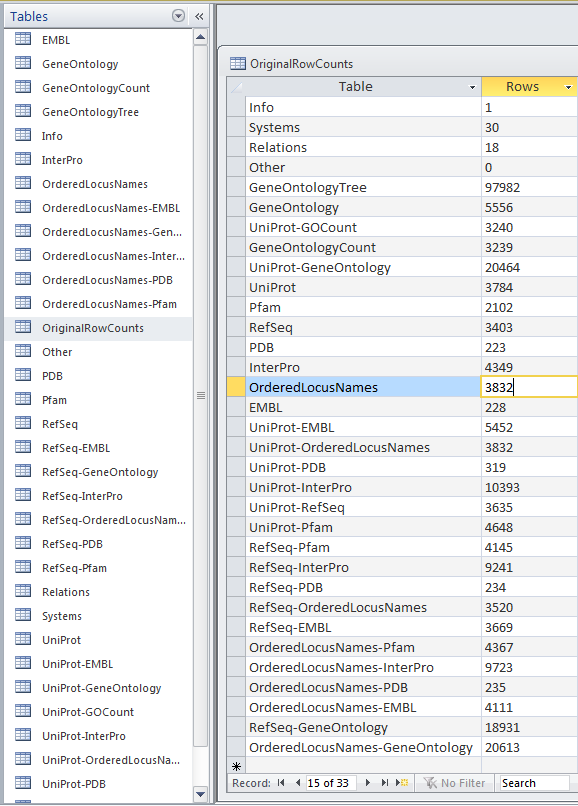
Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
Benchmark .gdb file: (for the Week 9 Assignment, use the "Vc-Std_External_20101022.gdb" as your benchmark, downloadable from here.
Copy the OriginalRowCounts table and paste it here:
- Vc-Std_20131022-HDelgadi-gmb2b70.gdb:
- Vc-Std_External_20101022.gdb:
Note: The results in Microsoft Access was 3832 Ordered Locus Name, but in the TallyEngine the results for Ordered Locus Name was 3831. The discrepancy is due to the proteins 1738 and 1739 were misinterpreted by Microsoft Access when they were inputted like 1738/1739 which was suppose to signify they are related, but instead this program viewed it as being separate proteins. Furthermore the difference between both Vc-Std_20131022-HDelgadi-gmb2b70.gdb and Vc-Std_External_20101022.gdb in Microsoft Access in terms of their OriginalRowCounts is that the benchmark, Vc-Std_External_20101022.gdb, includes more rows with table names such as: UniProt-Geneld RefSeq-Geneld Geneld-Pfam Geneld-InterPro Geneld-PDB Geneld-OrderedLocusNames Geneld-EMBL
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
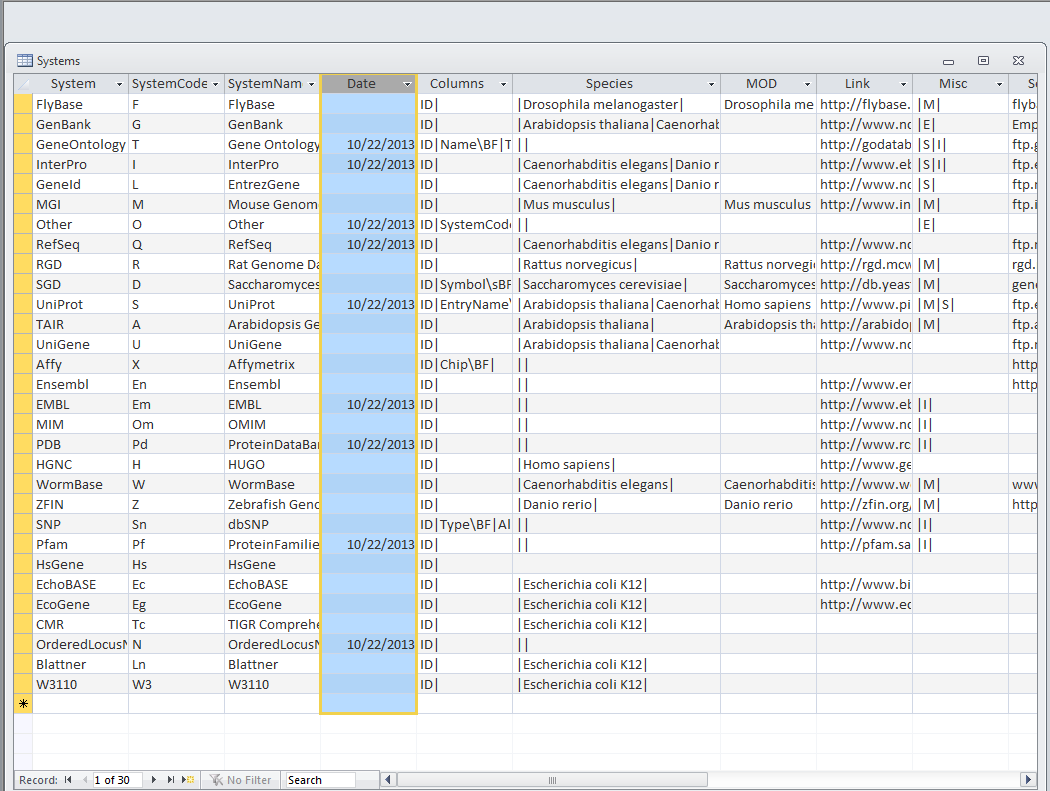
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
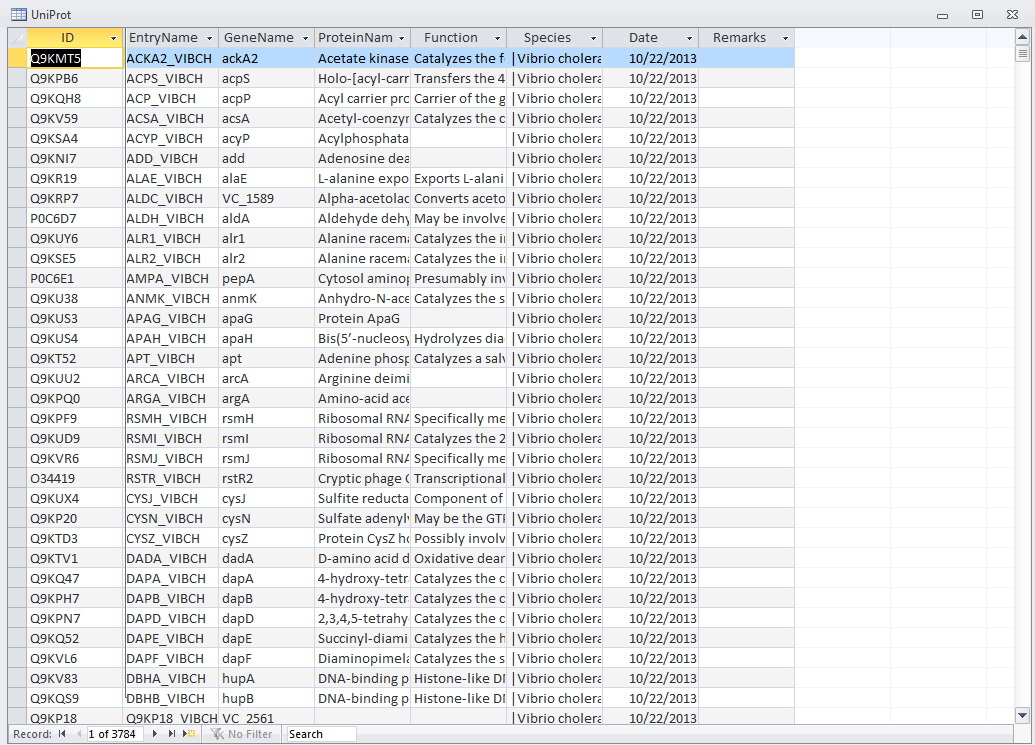
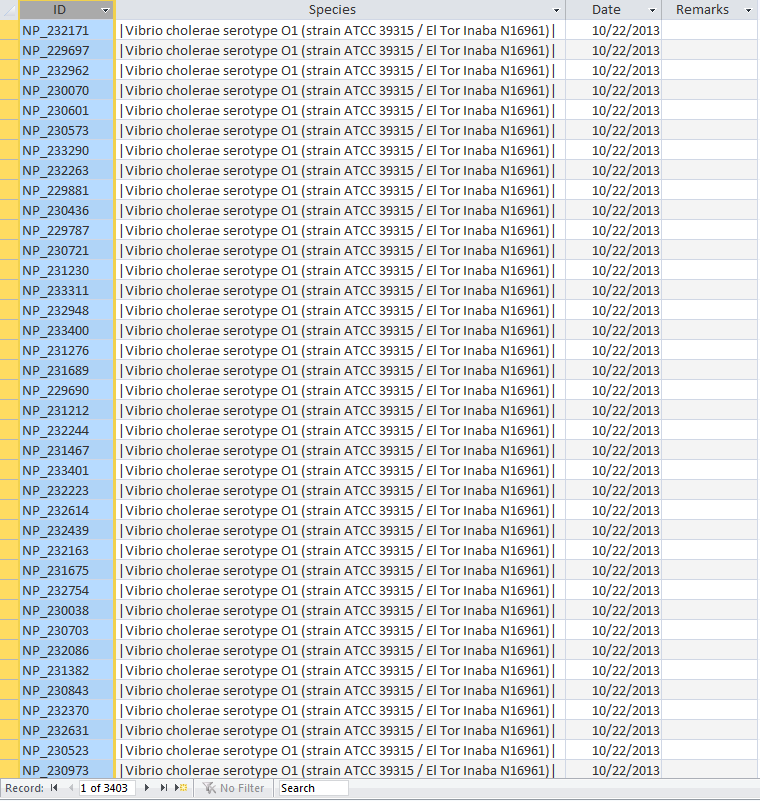
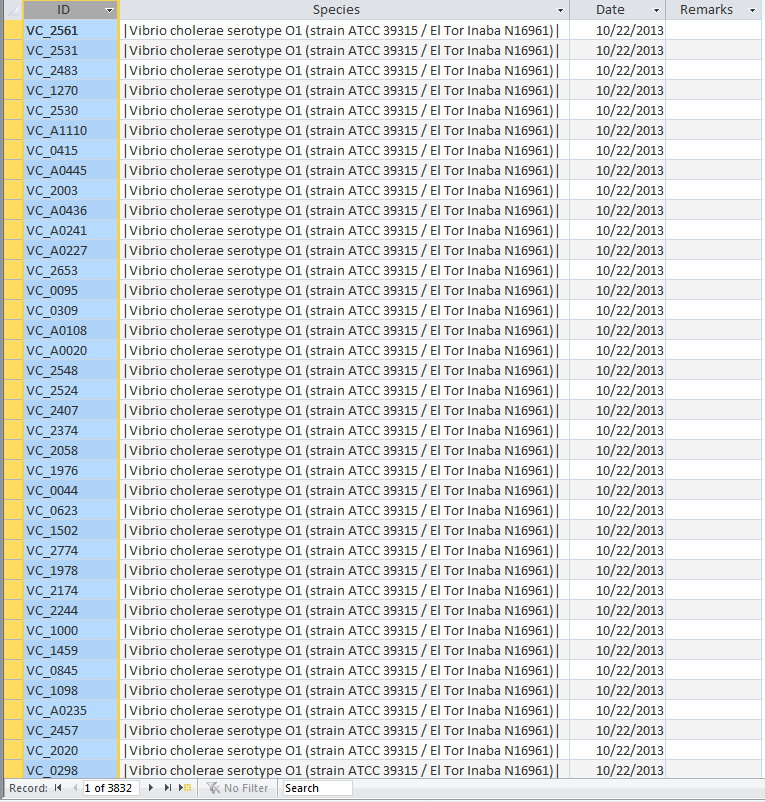
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
Note:
- There doesn't appear to be a set date in the Date field for all the gene ID systems present in the database.
- The IDs actually look very different from one another while RefSeq and OrderedLocusNames have underscores and UniProt does not.
UniProt
.gdb Use in GenMAPP
Note: We cannot input the IDs into GenMAPP since GenMAPP won't recognize IDs with the underscores.
Putting a gene on the MAPP using the GeneFinder window
- Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
Note:
Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
Note:
Coloring a MAPP with expression data
Note:
Running MAPPFinder
Note:
Compare Gene Database to Outside Resource
The OrderedLocusNames IDs in the exported Gene Database are derived from the UniProt XML. It is a good idea to check your list of OrderedLocusNames IDs to see how complete it is using the original source of the data (the sequencing organization, the MOD, etc.) Because UniProt is a protein database, it does not reference any non-protein genome features such as genes that code for functional RNAs, centromeres, telomeres, etc.
Note:
HDelgadi (talk) 13:37, 24 October 2013 (PDT)
- Link: [Protegen]
HDelgadi Week 9