HDelgadi Week 5
UniProt Exercise
In order to search for the ID P00533, which is known as the primary accession number and looks is used to search for the corresponding protein in the database, I used the following URL: http://www.uniprot.org/. I typed in the given ID number, P00533, under the Query subheading at the top near the "Search" button to its right. The search button was clicked on and the following information about this ID number which stands for the protein of interest was shown,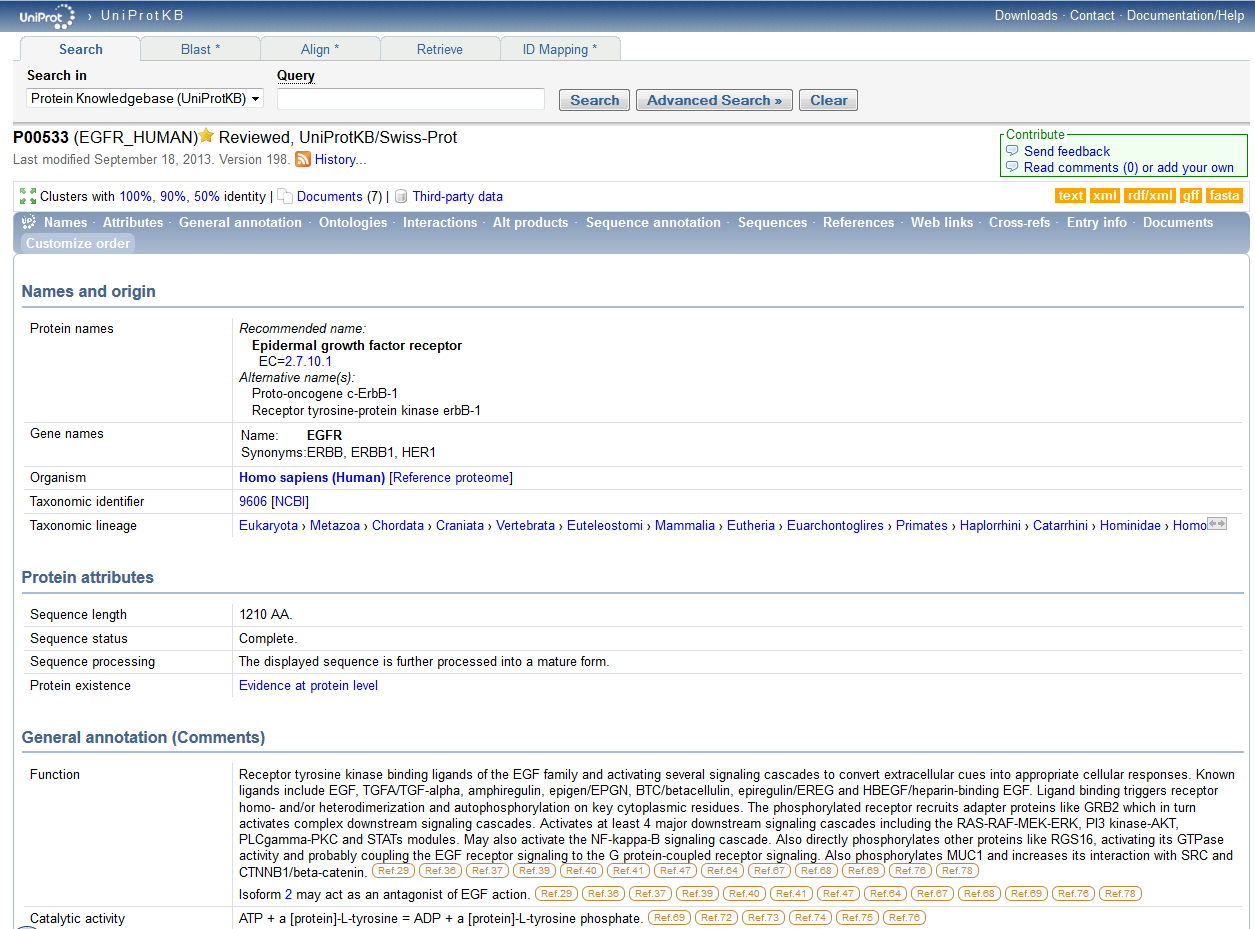
In order to get to the "ENZYME entry: EC 2.7.10.1" page which is referred to as the NiceZyme view of the enzyme in the reading, I had to click on EC=2.7.10.1 on the right of "Protein names" and under Recommended name. The subsequent page is the one as shown, 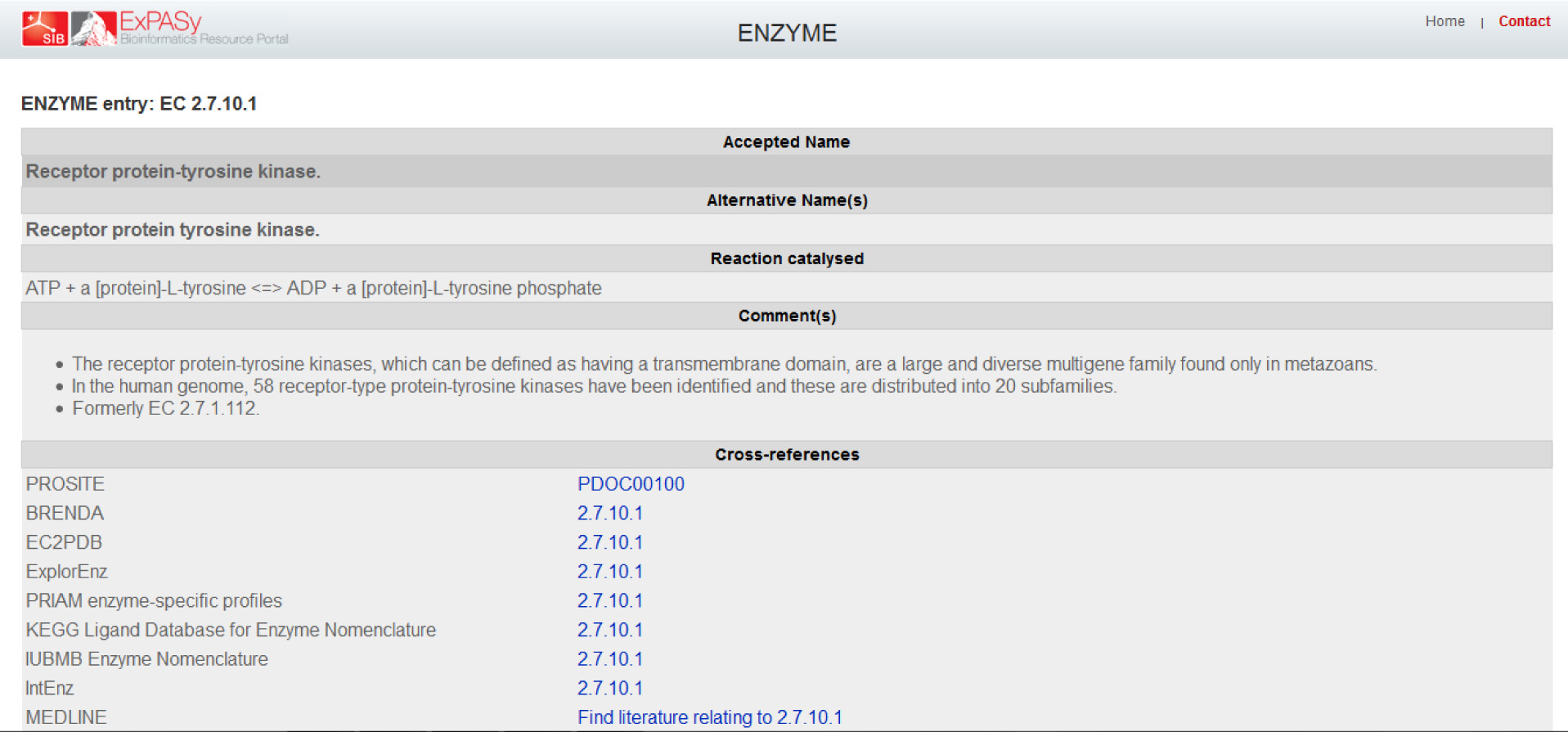
Furthermore, if I click on 9606 that is positioned to the right of "Taxonomic identifier", I can assess the taxonomy of where the protein is found, in this case in Humans. The page after clicking the Taxonomic identifier should look like this,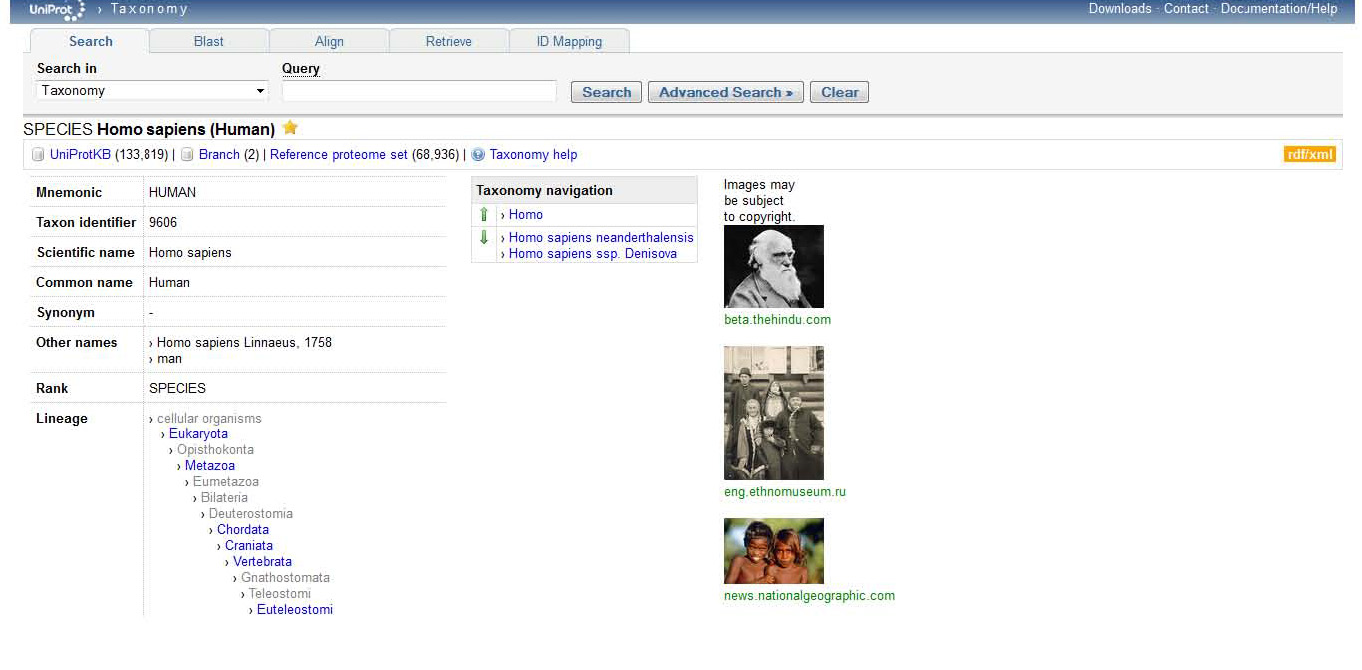
When exploring the comment section, I clicked on "General annotation (Comments)" heading in the light blue color. This took me to a "Help" box which gives you a one sentence brief description of what this section is about. Then I clicked on "More..." highlighted in a bright purple color which then led me to the "General annotation (Comments)" page with a subsection and content. This page looks like this: 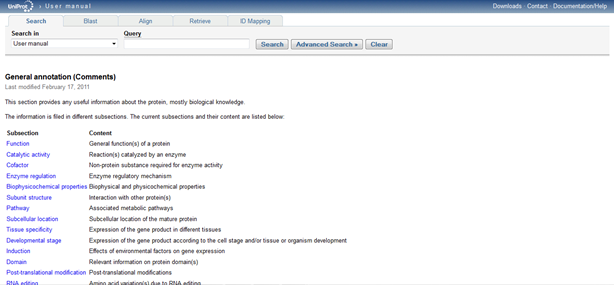 .
.
I clicked on X00588 mRNA. Translation: CAA25240.1. under "Cross-references" while EMBL is marked off. This sent me to the European Nucleotide Archive which gives you further information about your protein in terms of its nucleotide sequence. The page looks like this, 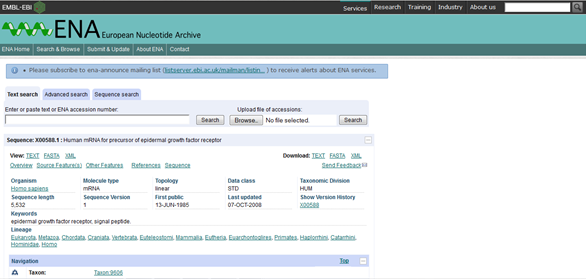 .
.
I then clicked on SWISS-2DPAGE under 2D gel databases which takes you to a "Help" box. I then clicked on the only URL that was in the box and the following information should appear,  .
.
I clicked on InterPro under "Family and domain databases". I clicked on Graphical view that is to the right column in terms of InterPro. This showed me the different domains, such as this, 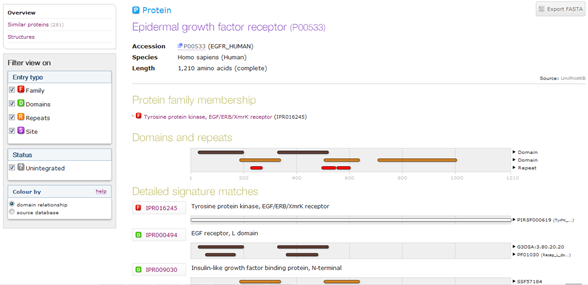 . Furthermore, clicking on InterPro directs me to a "Help" box and clicking on the only URL takes me to an overview of the protein information which looks like this,
. Furthermore, clicking on InterPro directs me to a "Help" box and clicking on the only URL takes me to an overview of the protein information which looks like this, 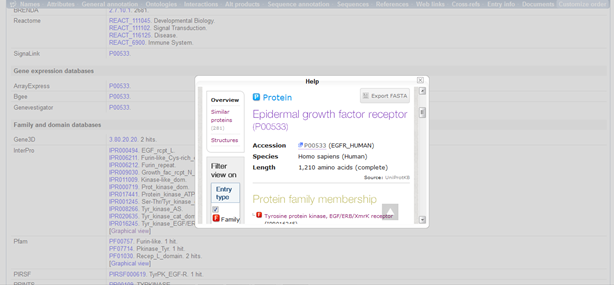 .
.
I clicked on Chain under the light blue subheading "Sequence annotation (Features)" which gives you a description of this subsection. The "Help" box will appear, but clicking the bright purple "More..." will take you to the desired page,  .
.
I clicked on the fasta button highlighted in yellow on the top far right hand side corner right below the "Contribute" box which is colored neon green. The protein sequence is then provided.  .
.
Write a one-paragraph summary of what you have learned about the human EGFR protein from this exercise.
I learned that the human EGFR protein is a receptor in triggering cell division, so it potentially triggers cancer. This particular protein is a protein kinase and its tissue specificity is in the placenta and in terms of ovarian cancer. A subcellular location is in the Endoplasmic reticulum membrane and its involvement in disease includes affecting tissue in the lung causing lung cancer.
Reflect and answer the following questions on your individual journal page:
- What was the purpose of this exercise?
The purpose of this exercise was to get familiar with the database UniProt and in doing so learning about the EGFR protein.
- What did I learn from this exercise?
I learned how to navigate my way through this database in hopes of finding further information and virtually all information about this protein.
- What did I not understand (yet) about this exercise?
I am still a bit confused about the various what seems like endless data under "Binary interactions" that I would like to further explore.
HDelgadi Week 5
- Link: [Protegen]